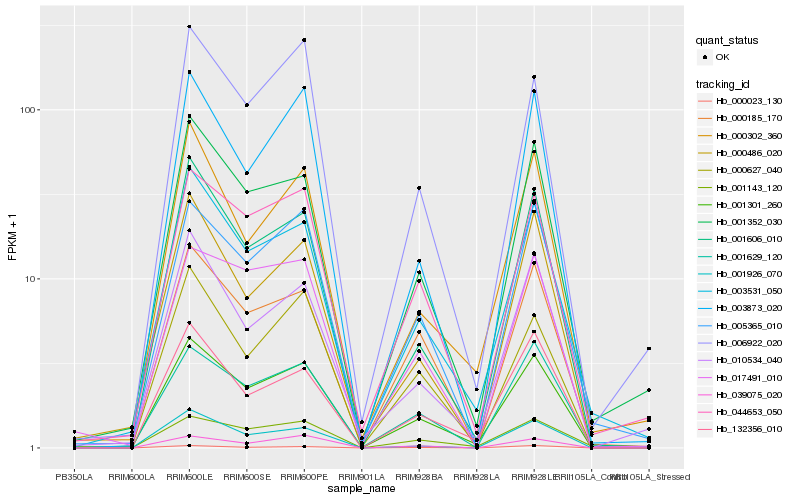
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001301_260 |
0.0 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001606_010 |
0.0883242664 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_003531_050 |
0.0893071154 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001352_030 |
0.0931415169 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 5 |
Hb_044653_050 |
0.0937125689 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001629_120 |
0.0979156705 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 7 |
Hb_132356_010 |
0.1000151759 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 8 |
Hb_005365_010 |
0.1001433222 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 9 |
Hb_000185_170 |
0.1027868902 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_000023_130 |
0.1049606759 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 11 |
Hb_001143_120 |
0.1059591906 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 12 |
Hb_003873_020 |
0.1076690321 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 13 |
Hb_000302_360 |
0.1090179568 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 14 |
Hb_000486_020 |
0.1098611661 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 15 |
Hb_039075_020 |
0.1104815965 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 16 |
Hb_000627_040 |
0.1113612612 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 17 |
Hb_010534_040 |
0.1129342789 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006922_020 |
0.1131403779 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 19 |
Hb_001926_070 |
0.1131986559 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 20 |
Hb_017491_010 |
0.114222583 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |