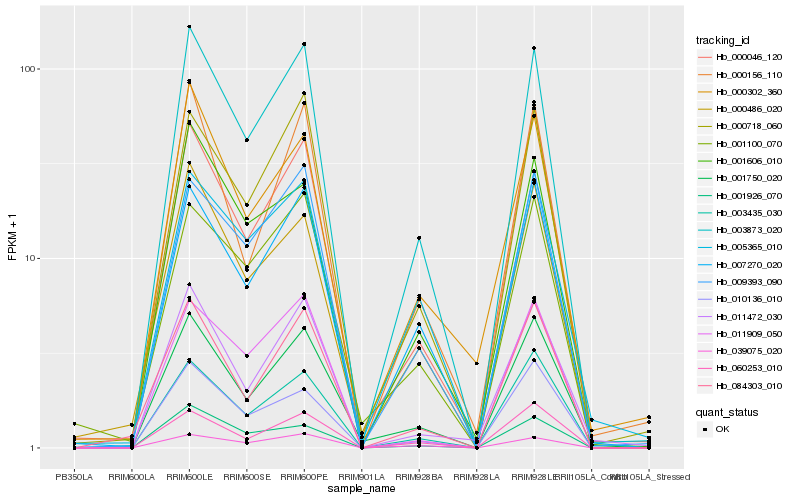
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003873_020 |
0.0 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 2 |
Hb_007270_020 |
0.0777742064 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 3 |
Hb_000718_060 |
0.0805048234 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_009393_090 |
0.0806291021 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_001750_020 |
0.0826541581 |
- |
- |
PREDICTED: MATE efflux family protein 6-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_039075_020 |
0.0843574381 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 7 |
Hb_003435_030 |
0.0882323745 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_000486_020 |
0.0984378184 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 9 |
Hb_011909_050 |
0.1004147592 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 10 |
Hb_001606_010 |
0.1005702389 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_060253_010 |
0.1010893625 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 12 |
Hb_084303_010 |
0.1012046847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000156_110 |
0.1012334272 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 14 |
Hb_011472_030 |
0.1015943393 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 15 |
Hb_001926_070 |
0.1048146667 |
- |
- |
hypothetical protein JCGZ_21510 [Jatropha curcas] |
| 16 |
Hb_000302_360 |
0.1052554669 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 17 |
Hb_010136_010 |
0.1059476369 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 18 |
Hb_005365_010 |
0.1063529316 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 19 |
Hb_000046_120 |
0.1068776939 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_001100_070 |
0.1076047099 |
- |
- |
PREDICTED: ferredoxin--nitrite reductase, chloroplastic isoform X1 [Jatropha curcas] |