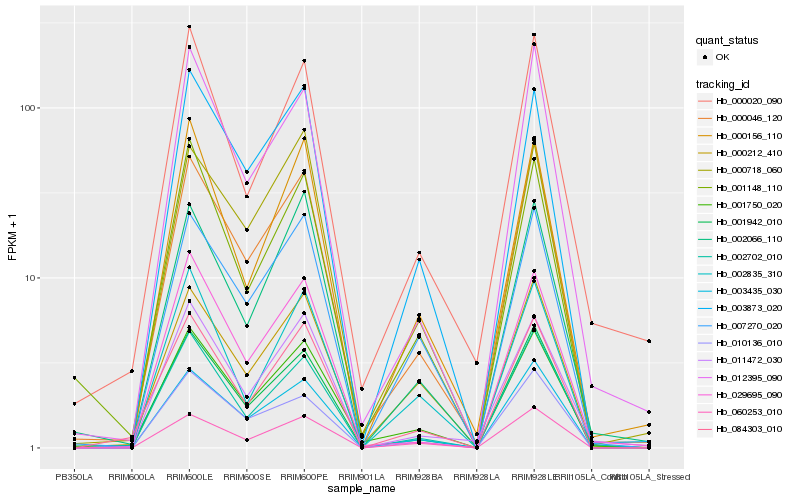
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001750_020 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 6-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000046_120 |
0.0806230744 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_003873_020 |
0.0826541581 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 4 |
Hb_003435_030 |
0.0839122428 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_060253_010 |
0.0879265525 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 6 |
Hb_007270_020 |
0.0902246266 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 7 |
Hb_002702_010 |
0.0909420586 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 8 |
Hb_000212_410 |
0.0935209601 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 9 |
Hb_001942_010 |
0.0956027447 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 10 |
Hb_000156_110 |
0.0998458147 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 11 |
Hb_084303_010 |
0.1031471476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011472_030 |
0.1036159837 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 13 |
Hb_000718_060 |
0.1037779226 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_012395_090 |
0.1053083335 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 15 |
Hb_029695_090 |
0.1054725765 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 16 |
Hb_001148_110 |
0.1072568192 |
- |
- |
- |
| 17 |
Hb_002835_310 |
0.1072570097 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_010136_010 |
0.1074778483 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 19 |
Hb_000020_090 |
0.1076537654 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 20 |
Hb_002066_110 |
0.1080721929 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |