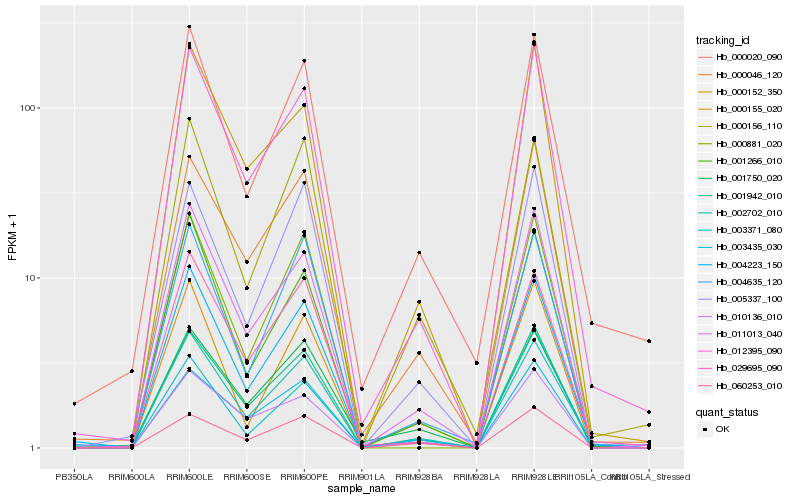
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002702_010 |
0.0 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 2 |
Hb_012395_090 |
0.050121799 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 3 |
Hb_011013_040 |
0.0653746633 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 4 |
Hb_004223_150 |
0.0786980388 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 5 |
Hb_000046_120 |
0.0789823016 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_001942_010 |
0.0796091232 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 7 |
Hb_003371_080 |
0.0801611198 |
- |
- |
hypothetical protein POPTR_0001s23660g [Populus trichocarpa] |
| 8 |
Hb_003435_030 |
0.0818642835 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_001266_010 |
0.0843460355 |
- |
- |
PREDICTED: uncharacterized protein LOC101206474 [Cucumis sativus] |
| 10 |
Hb_000155_020 |
0.0844329776 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000020_090 |
0.0846396864 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 12 |
Hb_010136_010 |
0.0903251196 |
- |
- |
Putative receptor-like protein kinase [Glycine soja] |
| 13 |
Hb_001750_020 |
0.0909420586 |
- |
- |
PREDICTED: MATE efflux family protein 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005337_100 |
0.0915032906 |
- |
- |
PREDICTED: uncharacterized protein LOC105629349 [Jatropha curcas] |
| 15 |
Hb_060253_010 |
0.0952387349 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 16 |
Hb_004635_120 |
0.0961411411 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 17 |
Hb_000152_350 |
0.097777706 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 18 |
Hb_029695_090 |
0.0980619631 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 19 |
Hb_000881_020 |
0.0986483799 |
- |
- |
hypothetical protein MIMGU_mgv1a022110mg, partial [Erythranthe guttata] |
| 20 |
Hb_000156_110 |
0.0992721699 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |