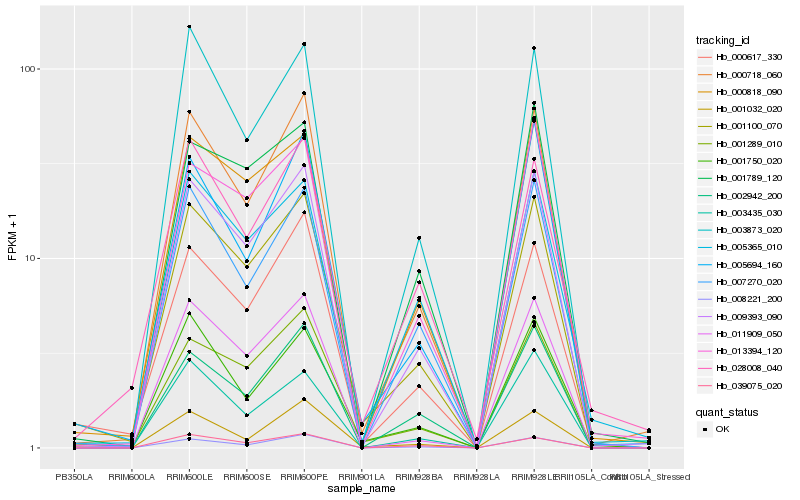
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001100_070 |
0.0 |
- |
- |
PREDICTED: ferredoxin--nitrite reductase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_001289_010 |
0.0577902754 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 3 |
Hb_009393_090 |
0.0697302749 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000818_090 |
0.0783363307 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 5 |
Hb_000718_060 |
0.0796501116 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000617_330 |
0.0839396172 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 7 |
Hb_007270_020 |
0.1002643594 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 8 |
Hb_005365_010 |
0.1041861162 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 9 |
Hb_011909_050 |
0.1049116799 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 10 |
Hb_013394_120 |
0.1056066795 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 11 |
Hb_008221_200 |
0.1066394703 |
- |
- |
PREDICTED: protein trichome birefringence-like 41 [Jatropha curcas] |
| 12 |
Hb_001789_120 |
0.1072884583 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_003873_020 |
0.1076047099 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 14 |
Hb_005694_160 |
0.1077979395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_039075_020 |
0.1079281315 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 16 |
Hb_002942_200 |
0.1153640957 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH137 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001750_020 |
0.1171957698 |
- |
- |
PREDICTED: MATE efflux family protein 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_028008_040 |
0.1172823135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001032_020 |
0.1189584988 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 20 |
Hb_003435_030 |
0.1216805555 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |