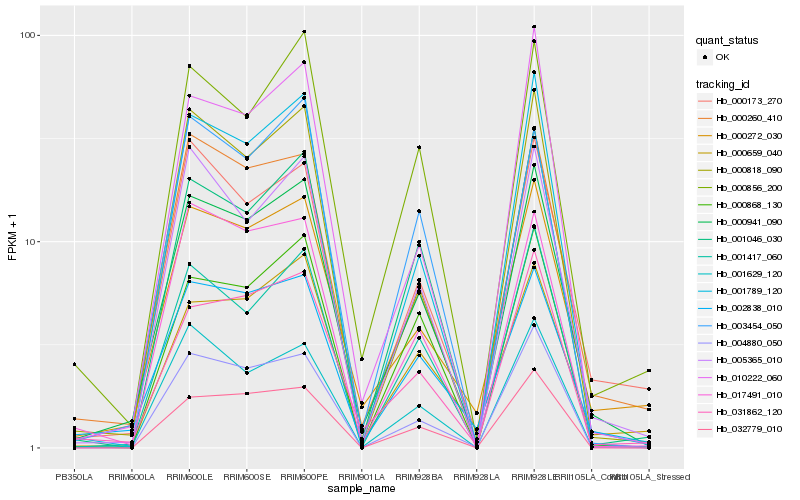
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000941_090 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 2 |
Hb_002838_010 |
0.0871834353 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 3 |
Hb_001417_060 |
0.0879815452 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 4 |
Hb_031862_120 |
0.0884415529 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 5 |
Hb_001789_120 |
0.0894263276 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000272_030 |
0.092588041 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000260_410 |
0.0932563574 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 8 |
Hb_005365_010 |
0.097252096 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 9 |
Hb_004880_050 |
0.0974242895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000856_200 |
0.0977401386 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 11 |
Hb_001046_030 |
0.1002676253 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000818_090 |
0.1009140705 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 13 |
Hb_001629_120 |
0.1013059374 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 14 |
Hb_000868_130 |
0.1051083839 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 15 |
Hb_000173_270 |
0.1061569147 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 16 |
Hb_032779_010 |
0.1078067325 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000659_040 |
0.1103055082 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_010222_060 |
0.1121429517 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_003454_050 |
0.11222714 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 20 |
Hb_017491_010 |
0.1124677394 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |