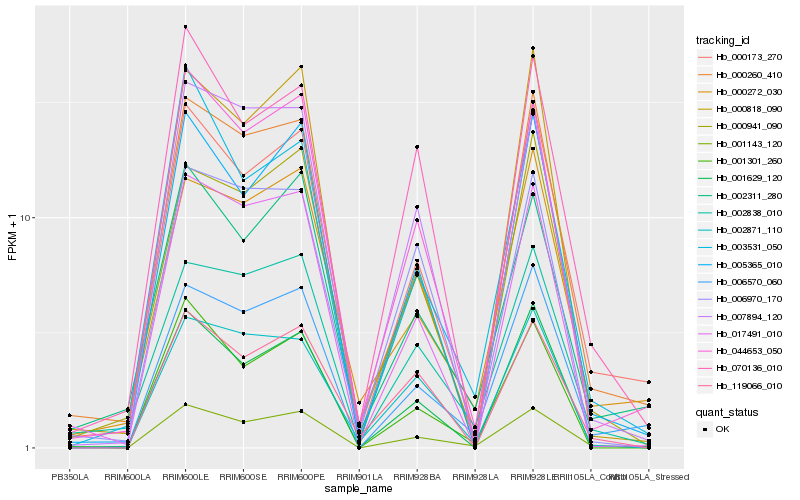
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001143_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 2 |
Hb_000173_270 |
0.1005184773 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001301_260 |
0.1059591906 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005365_010 |
0.1073966278 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 5 |
Hb_044653_050 |
0.1076064372 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002838_010 |
0.1084325575 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 7 |
Hb_000260_410 |
0.1110580146 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 8 |
Hb_006570_060 |
0.1133340996 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 9 |
Hb_017491_010 |
0.1174619998 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000941_090 |
0.118818161 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 11 |
Hb_001629_120 |
0.1192879606 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 12 |
Hb_002311_280 |
0.1222405726 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 13 |
Hb_007894_120 |
0.1263043502 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_070136_010 |
0.1265758681 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 15 |
Hb_003531_050 |
0.127854678 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002871_110 |
0.1294510017 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 17 |
Hb_119066_010 |
0.1308703152 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 18 |
Hb_000272_030 |
0.1320742068 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006970_170 |
0.133268282 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000818_090 |
0.1346238839 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |