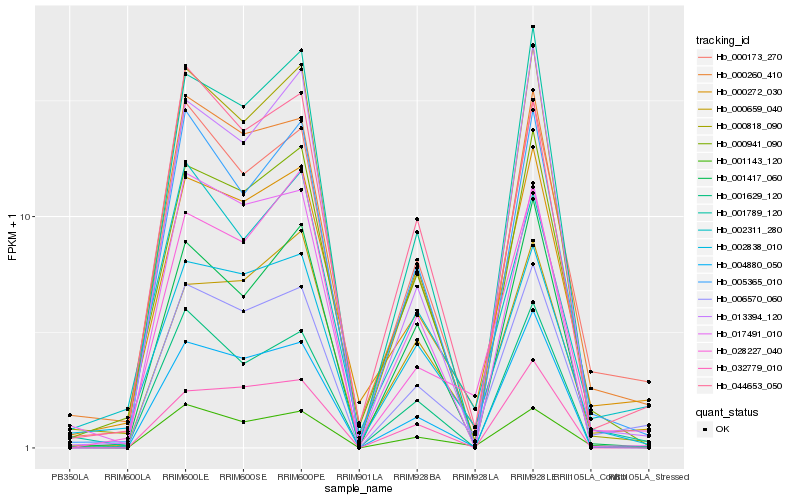
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006570_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 2 |
Hb_000173_270 |
0.0814926605 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000272_030 |
0.0881940508 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000260_410 |
0.0996454815 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 5 |
Hb_001143_120 |
0.1133340996 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 6 |
Hb_004880_050 |
0.1180517056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000659_040 |
0.1181804393 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000941_090 |
0.119537316 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 9 |
Hb_028227_040 |
0.1207783064 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 10 |
Hb_005365_010 |
0.1228865445 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 11 |
Hb_002838_010 |
0.1233748177 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 12 |
Hb_001629_120 |
0.1242138884 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 13 |
Hb_001789_120 |
0.1267437546 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_044653_050 |
0.1272806117 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000818_090 |
0.1298615786 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 16 |
Hb_002311_280 |
0.1340464917 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 17 |
Hb_013394_120 |
0.1397720234 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 18 |
Hb_001417_060 |
0.1405775594 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 19 |
Hb_017491_010 |
0.1406534545 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_032779_010 |
0.1423072611 |
- |
- |
unnamed protein product [Vitis vinifera] |