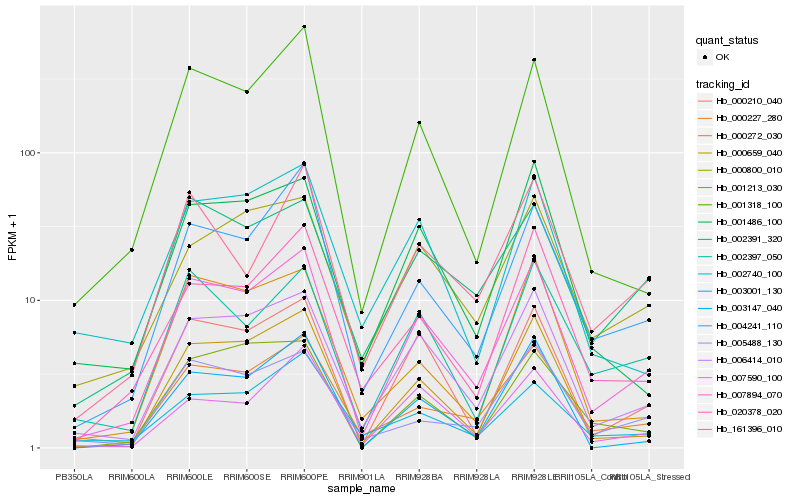
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 2 |
Hb_006414_010 |
0.0784772824 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 3 |
Hb_000210_040 |
0.1000135659 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 4 |
Hb_000227_280 |
0.1051828152 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 5 |
Hb_000659_040 |
0.1094705184 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004241_110 |
0.1164141977 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 7 |
Hb_001318_100 |
0.1196249031 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 8 |
Hb_020378_020 |
0.1298001684 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_005488_130 |
0.1327966175 |
- |
- |
transporter, putative [Ricinus communis] |
| 10 |
Hb_001486_100 |
0.1333883608 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 11 |
Hb_002391_320 |
0.1342091047 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 12 |
Hb_000800_010 |
0.1344415795 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 13 |
Hb_002397_050 |
0.1362246105 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007590_100 |
0.1380421324 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 15 |
Hb_003147_040 |
0.13888811 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 16 |
Hb_003001_130 |
0.1400000017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_161396_010 |
0.1434782439 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 18 |
Hb_002740_100 |
0.1435011142 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 19 |
Hb_000272_030 |
0.1441780571 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001213_030 |
0.1465234592 |
- |
- |
PAR1 protein [Theobroma cacao] |