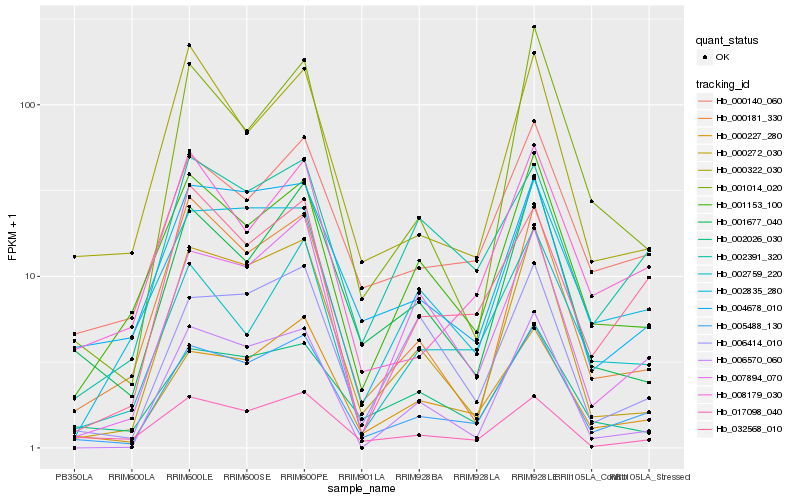
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005488_130 |
0.0 |
- |
- |
transporter, putative [Ricinus communis] |
| 2 |
Hb_000227_280 |
0.0876576462 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 3 |
Hb_000140_060 |
0.1192669935 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 4 |
Hb_000272_030 |
0.122494908 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004678_010 |
0.1260550207 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 6 |
Hb_007894_070 |
0.1327966175 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 7 |
Hb_001153_100 |
0.1332504331 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 8 |
Hb_002835_280 |
0.1371756331 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 9 |
Hb_000181_330 |
0.1378177224 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 10 |
Hb_032568_010 |
0.1385900045 |
- |
- |
PREDICTED: protein phosphatase 2C 56-like [Jatropha curcas] |
| 11 |
Hb_001677_040 |
0.1391447536 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 12 |
Hb_008179_030 |
0.1428246618 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 8 [Jatropha curcas] |
| 13 |
Hb_002391_320 |
0.146130225 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 14 |
Hb_002026_030 |
0.146676254 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 15 |
Hb_000322_030 |
0.1476055376 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 16 |
Hb_006570_060 |
0.1477353842 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 17 |
Hb_017098_040 |
0.1482315737 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 18 |
Hb_002759_220 |
0.148833011 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 19 |
Hb_006414_010 |
0.1490105079 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 20 |
Hb_001014_020 |
0.149119902 |
- |
- |
gene GA family protein [Populus trichocarpa] |