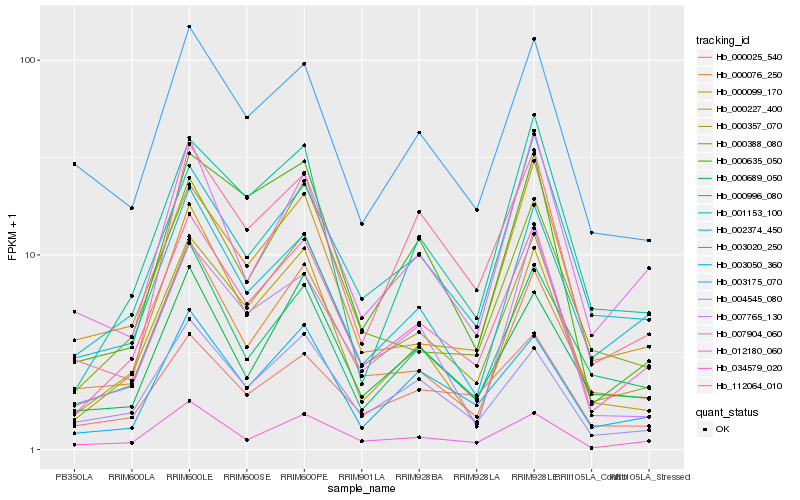
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034579_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000357_070 |
0.0643463268 |
- |
- |
- |
| 3 |
Hb_012180_060 |
0.0907093785 |
- |
- |
peptidase, putative [Ricinus communis] |
| 4 |
Hb_003050_360 |
0.1081239235 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004545_080 |
0.1102895097 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_002374_450 |
0.1122458849 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003020_250 |
0.1157068641 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 8 |
Hb_000996_080 |
0.1171768708 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 9 |
Hb_112064_010 |
0.1189345124 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 10 |
Hb_000076_250 |
0.1192019472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000388_080 |
0.120254365 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 12 |
Hb_000635_050 |
0.1206088068 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 13 |
Hb_001153_100 |
0.1208925862 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 14 |
Hb_007765_130 |
0.1230219598 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000099_170 |
0.1233798328 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 16 |
Hb_003175_070 |
0.124316906 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 17 |
Hb_000689_050 |
0.1246141617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000025_540 |
0.12470946 |
- |
- |
PREDICTED: uncharacterized protein LOC104879644 [Vitis vinifera] |
| 19 |
Hb_000227_400 |
0.1269647134 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 20 |
Hb_007904_060 |
0.1275920321 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |