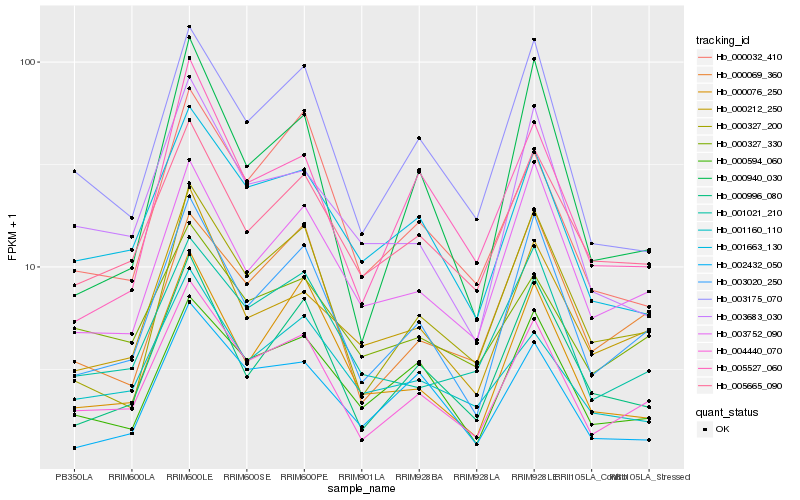
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004440_070 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520 [Jatropha curcas] |
| 2 |
Hb_003020_250 |
0.079013662 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 3 |
Hb_001663_130 |
0.1072829584 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 4 |
Hb_003175_070 |
0.1073639791 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 5 |
Hb_000327_200 |
0.1123314397 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 6 |
Hb_001021_210 |
0.1144911827 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005665_090 |
0.1165327802 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 8 |
Hb_000594_060 |
0.1192478977 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 9 |
Hb_000996_080 |
0.1195328862 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 10 |
Hb_003752_090 |
0.1230042127 |
- |
- |
chitinase, putative [Ricinus communis] |
| 11 |
Hb_000212_250 |
0.1232967447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000032_410 |
0.1244646245 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 13 |
Hb_001160_110 |
0.1265864807 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 14 |
Hb_000327_330 |
0.1272869227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000069_360 |
0.1272908477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000076_250 |
0.1290824503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003683_030 |
0.1299579702 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 18 |
Hb_000940_030 |
0.130676336 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005527_060 |
0.1307578015 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 20 |
Hb_002432_050 |
0.1327023146 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |