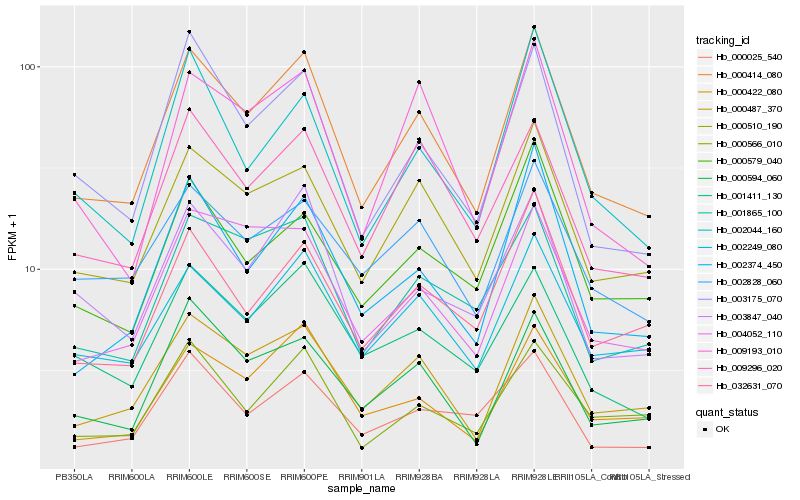
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000414_080 |
0.0 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000487_370 |
0.0660304612 |
- |
- |
- |
| 3 |
Hb_000510_190 |
0.0734281558 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 4 |
Hb_002249_080 |
0.0867756535 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 5 |
Hb_000422_080 |
0.0928964066 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 6 |
Hb_003175_070 |
0.0929516222 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 7 |
Hb_002374_450 |
0.0946631747 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000594_060 |
0.0961824261 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 9 |
Hb_001865_100 |
0.0968350299 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 10 |
Hb_032631_070 |
0.0981490703 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_003847_040 |
0.098536796 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 12 |
Hb_009296_020 |
0.1010385934 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001411_130 |
0.1016213914 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002044_160 |
0.1021436228 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 15 |
Hb_009193_010 |
0.1024085912 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 16 |
Hb_004052_110 |
0.1024639124 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000566_010 |
0.1041647382 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 18 |
Hb_002828_060 |
0.105095546 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000579_040 |
0.1062086864 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 20 |
Hb_000025_540 |
0.1063802591 |
- |
- |
PREDICTED: uncharacterized protein LOC104879644 [Vitis vinifera] |