| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
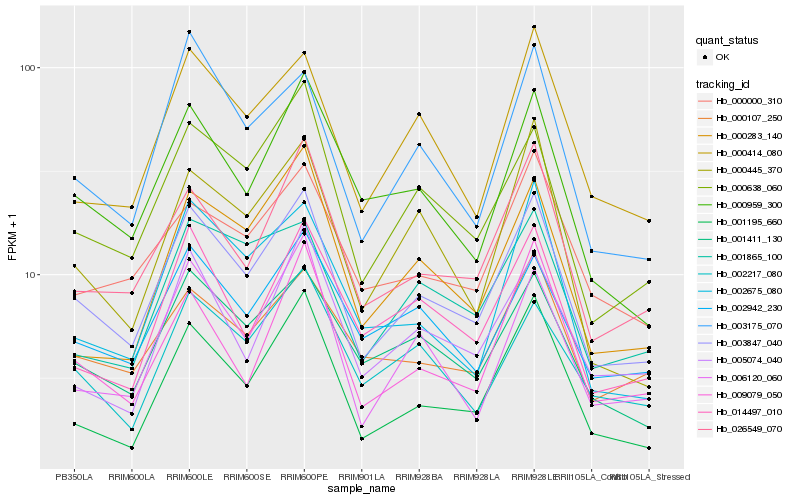
Hb_003847_040 |
0.0 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 2 |
Hb_000638_060 |
0.0870151523 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 3 |
Hb_001411_130 |
0.0887676427 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 4 |
Hb_026549_070 |
0.0892505844 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 5 |
Hb_000959_300 |
0.0964304067 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 6 |
Hb_001195_660 |
0.0964406301 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 7 |
Hb_000414_080 |
0.098536796 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002942_230 |
0.0987507884 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 9 |
Hb_003175_070 |
0.1020504709 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 10 |
Hb_000107_250 |
0.1064557452 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 11 |
Hb_014497_010 |
0.1069533561 |
- |
- |
PREDICTED: uncharacterized protein LOC105635153 [Jatropha curcas] |
| 12 |
Hb_000000_310 |
0.1094087253 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 13 |
Hb_002217_080 |
0.1096393807 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 16 [Jatropha curcas] |
| 14 |
Hb_006120_060 |
0.1100519958 |
- |
- |
O-acetyltransferase family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_009079_050 |
0.1127181135 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 16 |
Hb_000445_370 |
0.1141078063 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 17 |
Hb_002675_080 |
0.1143127254 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 18 |
Hb_001865_100 |
0.1144686945 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 19 |
Hb_000283_140 |
0.1163607611 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_005074_040 |
0.1166796609 |
- |
- |
ATP binding protein, putative [Ricinus communis] |