| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
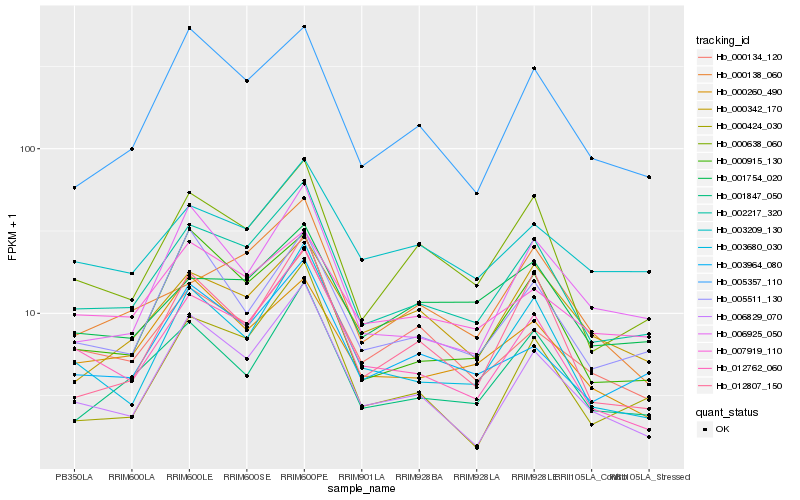
Hb_002217_320 |
0.0 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_012762_060 |
0.0863131044 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_000638_060 |
0.0887321269 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 4 |
Hb_001847_050 |
0.0947631035 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 5 |
Hb_003964_080 |
0.1004352484 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 6 |
Hb_003680_030 |
0.1026400497 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 7 |
Hb_006829_070 |
0.1043445375 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 8 |
Hb_000134_120 |
0.1088468622 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 9 |
Hb_005357_110 |
0.1118009854 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 10 |
Hb_000138_060 |
0.115184447 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 11 |
Hb_012807_150 |
0.1169895982 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 12 |
Hb_000915_130 |
0.1175686117 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 13 |
Hb_003209_130 |
0.1183386944 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 14 |
Hb_005511_130 |
0.1201155035 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 15 |
Hb_006925_050 |
0.1203402039 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 16 |
Hb_007919_110 |
0.1209549749 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 17 |
Hb_000260_490 |
0.1226152531 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 18 |
Hb_001754_020 |
0.1226631211 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 19 |
Hb_000342_170 |
0.1227270664 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 20 |
Hb_000424_030 |
0.1240317814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |