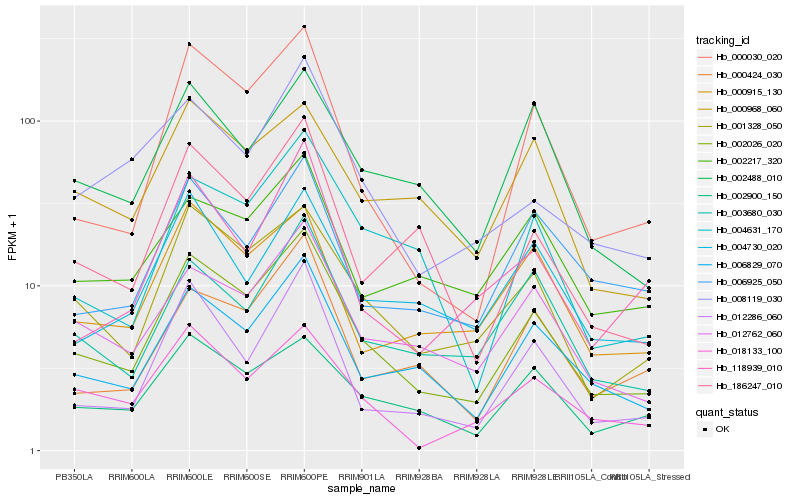
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002026_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 2 |
Hb_001328_050 |
0.1009417658 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 3 |
Hb_006829_070 |
0.1024965765 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 4 |
Hb_000030_020 |
0.1131513226 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 5 |
Hb_012762_060 |
0.1160173817 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_002900_150 |
0.1268364745 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 7 |
Hb_003680_030 |
0.1302078509 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 8 |
Hb_004631_170 |
0.1321044691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_012286_060 |
0.1328401998 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000424_030 |
0.1339358617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_018133_100 |
0.1350987356 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_186247_010 |
0.1357663117 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 13 |
Hb_000915_130 |
0.1385726756 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 14 |
Hb_008119_030 |
0.1401559442 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 15 |
Hb_002488_010 |
0.1405922336 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 16 |
Hb_002217_320 |
0.1427356117 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006925_050 |
0.1427793512 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 18 |
Hb_004730_020 |
0.1455631811 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 19 |
Hb_000968_060 |
0.1457119802 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 20 |
Hb_118939_010 |
0.150528814 |
- |
- |
protein binding protein, putative [Ricinus communis] |