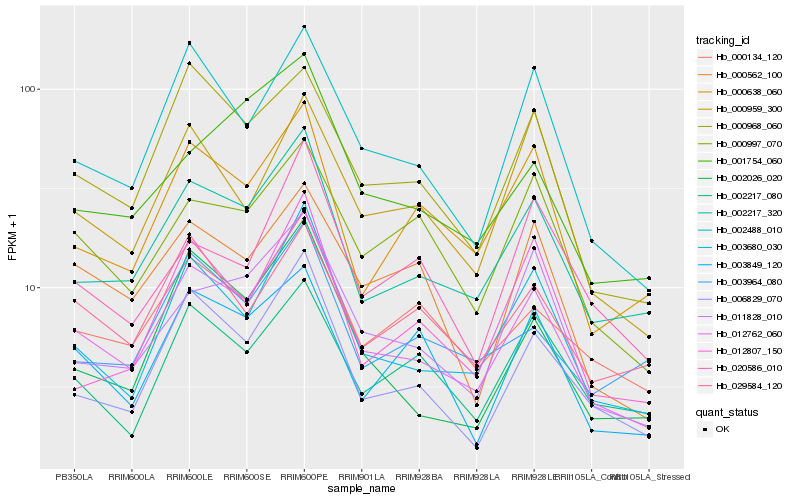
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012762_060 |
0.0 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_003680_030 |
0.0770278671 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 3 |
Hb_002217_320 |
0.0863131044 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006829_070 |
0.0914275005 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 5 |
Hb_002488_010 |
0.1023492861 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 6 |
Hb_002026_020 |
0.1160173817 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 7 |
Hb_000134_120 |
0.1161396491 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 8 |
Hb_000968_060 |
0.1172776741 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 9 |
Hb_011828_010 |
0.1238198927 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 10 |
Hb_000638_060 |
0.1280899308 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 11 |
Hb_000997_070 |
0.1308013989 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 12 |
Hb_000959_300 |
0.1336392191 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 13 |
Hb_000562_100 |
0.1357450627 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 14 |
Hb_029584_120 |
0.1371715992 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_012807_150 |
0.1380307126 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 16 |
Hb_001754_060 |
0.1385624976 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_003964_080 |
0.1390715171 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 18 |
Hb_003849_120 |
0.1394793195 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 19 |
Hb_020586_010 |
0.1396186884 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_002217_080 |
0.1396704559 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 16 [Jatropha curcas] |