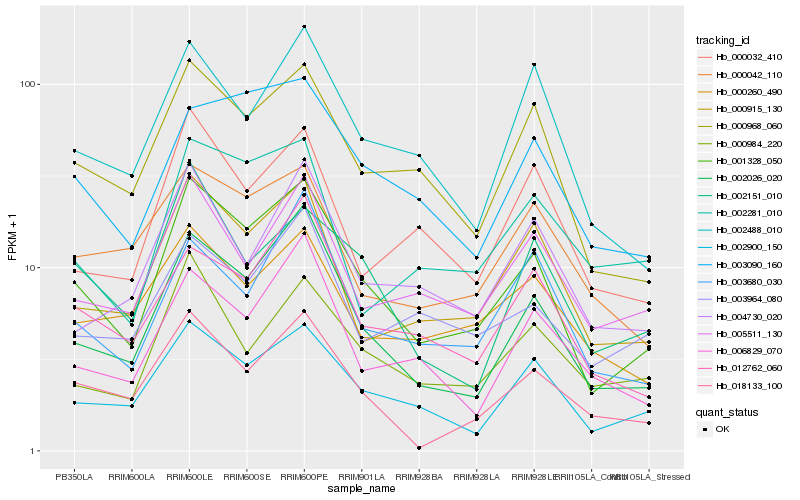
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001328_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 2 |
Hb_002026_020 |
0.1009417658 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 3 |
Hb_002900_150 |
0.1136061554 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 4 |
Hb_000968_060 |
0.1169118381 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 5 |
Hb_000915_130 |
0.1282805678 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 6 |
Hb_018133_100 |
0.1399203733 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_003090_160 |
0.1448024932 |
- |
- |
- |
| 8 |
Hb_012762_060 |
0.1451269649 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 9 |
Hb_005511_130 |
0.1462009839 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 10 |
Hb_002488_010 |
0.1484780174 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 11 |
Hb_000984_220 |
0.1511532226 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 12 |
Hb_002151_010 |
0.1519307927 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 13 |
Hb_000032_410 |
0.1541029286 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 14 |
Hb_000260_490 |
0.1554699302 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 15 |
Hb_003680_030 |
0.1571238808 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000042_110 |
0.1576447117 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 17 |
Hb_004730_020 |
0.1590646986 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 18 |
Hb_006829_070 |
0.1591223299 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 19 |
Hb_003964_080 |
0.1615701129 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 20 |
Hb_002281_010 |
0.1633383565 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |