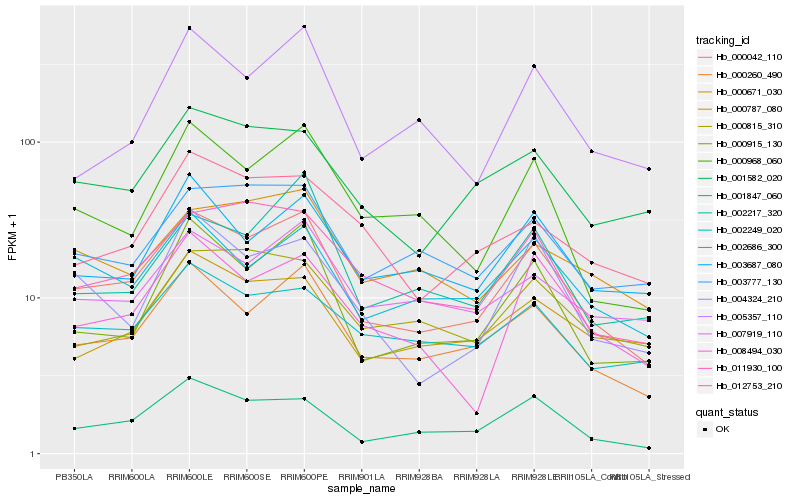
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_110 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 2 |
Hb_000260_490 |
0.0788240409 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 3 |
Hb_001847_060 |
0.0978292062 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Vitis vinifera] |
| 4 |
Hb_000915_130 |
0.1063548059 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 5 |
Hb_011930_100 |
0.1122286762 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 6 |
Hb_000968_060 |
0.1161105968 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 7 |
Hb_001582_020 |
0.1221573234 |
- |
- |
plastid acyl-ACP thioesterase [Vernicia fordii] |
| 8 |
Hb_003777_130 |
0.123420606 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 9 |
Hb_002686_300 |
0.1237870817 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 10 |
Hb_003687_080 |
0.1244453068 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 11 |
Hb_007919_110 |
0.1258484593 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 12 |
Hb_002217_320 |
0.127259197 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_008494_030 |
0.1282723314 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_002249_020 |
0.1292285631 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 15 |
Hb_004324_210 |
0.1300989058 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 16 |
Hb_005357_110 |
0.1310055263 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 17 |
Hb_000671_030 |
0.1315520068 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 18 |
Hb_000787_080 |
0.132218765 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 19 |
Hb_012753_210 |
0.1347539513 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 20 |
Hb_000815_310 |
0.1365155762 |
- |
- |
protein kinase, putative [Ricinus communis] |