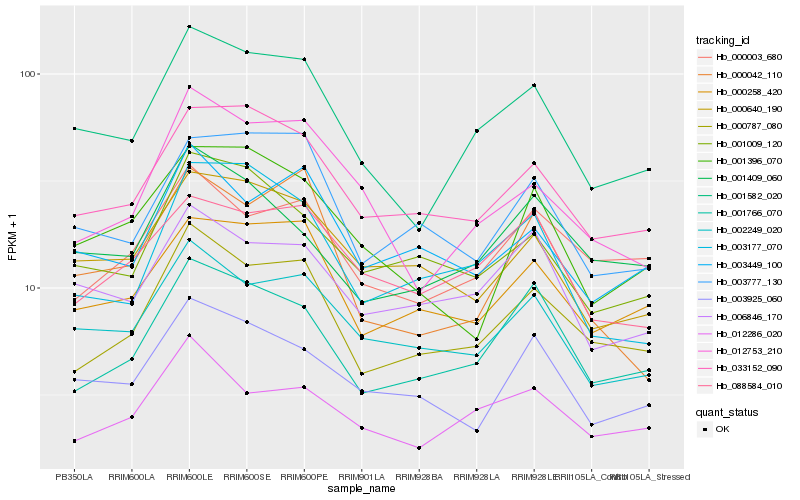
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001582_020 |
0.0 |
- |
- |
plastid acyl-ACP thioesterase [Vernicia fordii] |
| 2 |
Hb_002249_020 |
0.0971342408 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 3 |
Hb_033152_090 |
0.0971998589 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 4 |
Hb_012753_210 |
0.0998660171 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 5 |
Hb_000787_080 |
0.1035729541 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 6 |
Hb_000258_420 |
0.10504727 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 7 |
Hb_006846_170 |
0.1057887293 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 8 |
Hb_001766_070 |
0.1092681919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_012286_020 |
0.1100559896 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 10 |
Hb_088584_010 |
0.1133421946 |
- |
- |
hypothetical protein POPTR_0016s08470g [Populus trichocarpa] |
| 11 |
Hb_000640_190 |
0.1149404775 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003177_070 |
0.1172956341 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 13 |
Hb_001409_060 |
0.1184295073 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 14 |
Hb_003777_130 |
0.1192843885 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 15 |
Hb_001009_120 |
0.1193980278 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003925_060 |
0.1197360532 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000003_680 |
0.1212081243 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 18 |
Hb_000042_110 |
0.1221573234 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 19 |
Hb_001396_070 |
0.1241251833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003449_100 |
0.1252387819 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |