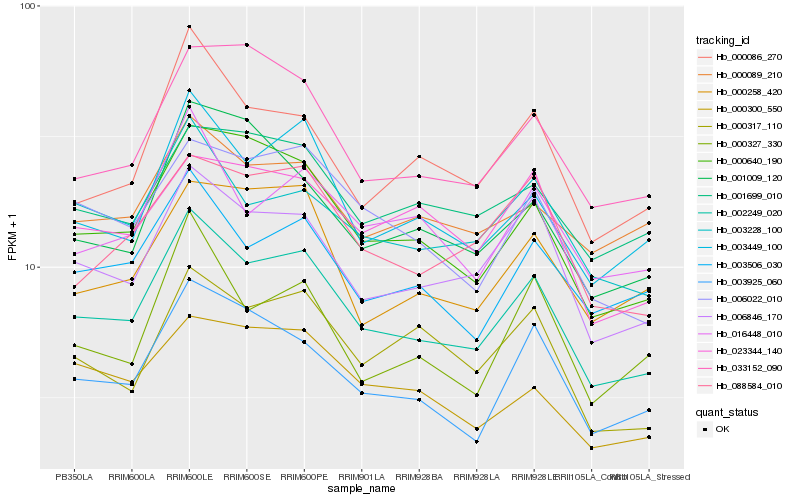
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002249_020 |
0.0 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 2 |
Hb_006846_170 |
0.0645393425 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 3 |
Hb_000640_190 |
0.0652419488 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003449_100 |
0.0718070186 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 5 |
Hb_003925_060 |
0.0772878784 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006022_010 |
0.0799908368 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 7 |
Hb_003228_100 |
0.0802609495 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 8 |
Hb_003506_030 |
0.0827012513 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_000086_270 |
0.0838650509 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 10 |
Hb_088584_010 |
0.0860915713 |
- |
- |
hypothetical protein POPTR_0016s08470g [Populus trichocarpa] |
| 11 |
Hb_016448_010 |
0.086789 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 12 |
Hb_033152_090 |
0.0871687984 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 13 |
Hb_000317_110 |
0.0901435017 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 14 |
Hb_000258_420 |
0.0921224473 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 15 |
Hb_000300_550 |
0.0923007644 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 16 |
Hb_000089_210 |
0.0931100015 |
- |
- |
unknown [Medicago truncatula] |
| 17 |
Hb_001699_010 |
0.0932498812 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 18 |
Hb_000327_330 |
0.0945130517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001009_120 |
0.0946473286 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 20 |
Hb_023344_140 |
0.0960665154 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |