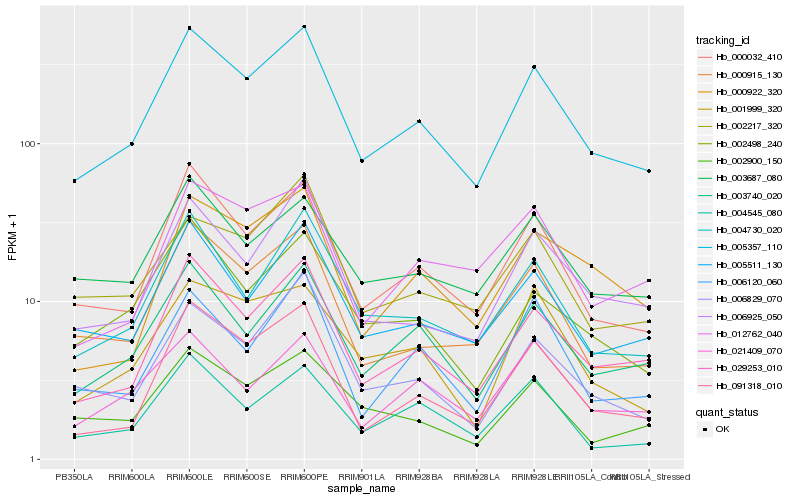
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005357_110 |
0.0 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 2 |
Hb_000032_410 |
0.0734365088 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 3 |
Hb_029253_010 |
0.0737548766 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 4 |
Hb_003740_020 |
0.0809811748 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_000915_130 |
0.0895034483 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 6 |
Hb_004730_020 |
0.0929679511 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 7 |
Hb_005511_130 |
0.0944870871 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 8 |
Hb_006925_050 |
0.100810004 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 9 |
Hb_091318_010 |
0.1065079109 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 10 |
Hb_003687_080 |
0.1083811971 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 11 |
Hb_002498_240 |
0.1084254245 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006829_070 |
0.1106233351 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 13 |
Hb_002217_320 |
0.1118009854 |
- |
- |
PREDICTED: probable O-acetyltransferase CAS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002900_150 |
0.1129193775 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 15 |
Hb_004545_080 |
0.1160580321 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_012762_040 |
0.116240929 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 17 |
Hb_001999_320 |
0.1165173461 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 18 |
Hb_021409_070 |
0.1168168868 |
- |
- |
PREDICTED: transcription factor bHLH157 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000922_320 |
0.1168784661 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 20 |
Hb_006120_060 |
0.1180951352 |
- |
- |
O-acetyltransferase family protein isoform 1 [Theobroma cacao] |