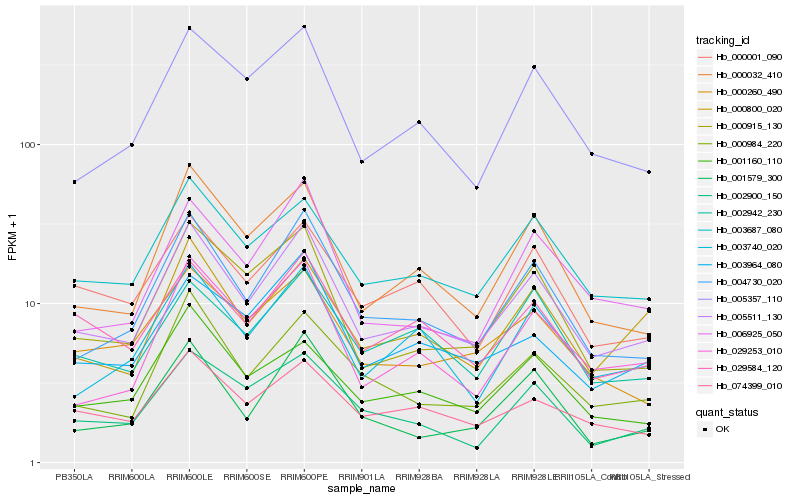
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005511_130 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 2 |
Hb_004730_020 |
0.0693290521 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 3 |
Hb_003687_080 |
0.0869436556 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 4 |
Hb_000032_410 |
0.0875342731 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 5 |
Hb_000984_220 |
0.0936524879 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 6 |
Hb_000915_130 |
0.09371733 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 7 |
Hb_005357_110 |
0.0944870871 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 8 |
Hb_029253_010 |
0.0971116032 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 9 |
Hb_003740_020 |
0.0985481429 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_006925_050 |
0.0992356958 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 11 |
Hb_074399_010 |
0.100781495 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 12 |
Hb_000800_020 |
0.1025162713 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 13 |
Hb_003964_080 |
0.1035832491 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 14 |
Hb_029584_120 |
0.1036446129 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000260_490 |
0.1045293729 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 16 |
Hb_001160_110 |
0.1076335587 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 17 |
Hb_000001_090 |
0.1079352293 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 18 |
Hb_001579_300 |
0.1098793045 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 19 |
Hb_002900_150 |
0.1101120972 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 20 |
Hb_002942_230 |
0.1115467285 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |