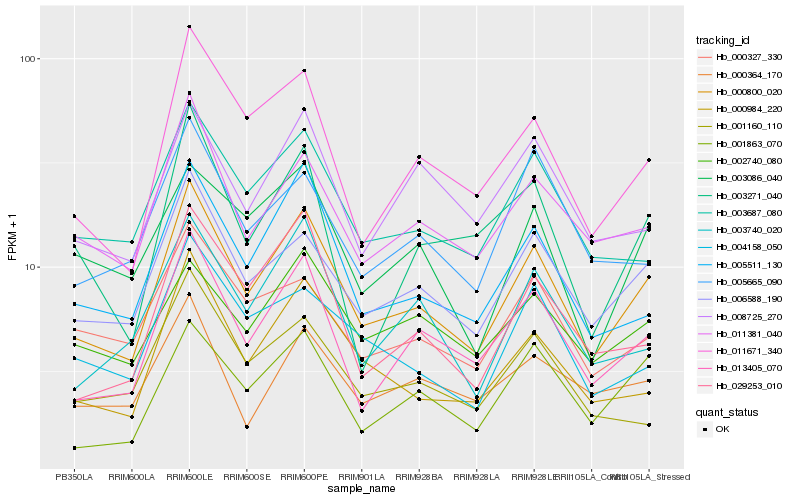
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000800_020 |
0.0 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 2 |
Hb_006588_190 |
0.086433143 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 3 |
Hb_013405_070 |
0.0884769206 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 4 |
Hb_011671_340 |
0.0989368696 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 5 |
Hb_005511_130 |
0.1025162713 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 6 |
Hb_011381_040 |
0.1031338028 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000984_220 |
0.1101250246 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 8 |
Hb_003687_080 |
0.1140277535 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 9 |
Hb_001863_070 |
0.114398192 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 10 |
Hb_029253_010 |
0.1211501524 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 11 |
Hb_003740_020 |
0.1211765779 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_000327_330 |
0.1241005252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004158_050 |
0.1248702587 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 14 |
Hb_002740_080 |
0.1275016424 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 15 |
Hb_008725_270 |
0.1279878141 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 16 |
Hb_003086_040 |
0.1293988218 |
- |
- |
PREDICTED: uncharacterized protein LOC105635780 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000364_170 |
0.1306002611 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 18 |
Hb_005665_090 |
0.1315796237 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 19 |
Hb_003271_040 |
0.1328187965 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 20 |
Hb_001160_110 |
0.1346209258 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |