| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
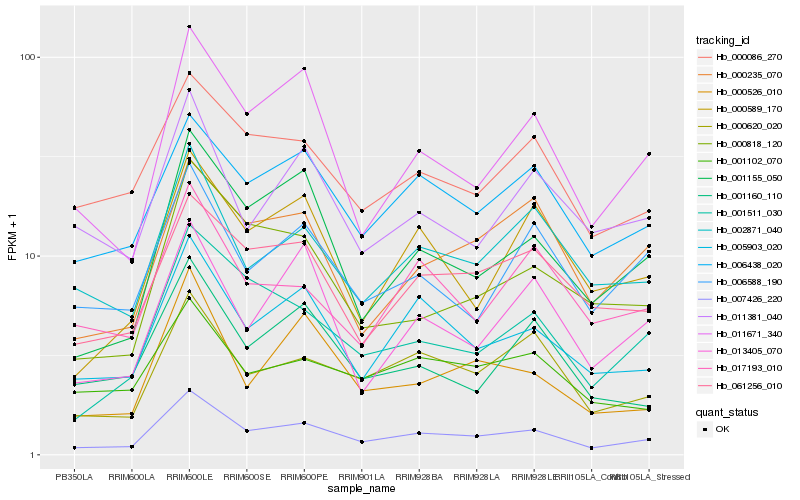
Hb_007426_220 |
0.0 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_005903_020 |
0.0998352878 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 3 |
Hb_011671_340 |
0.1078715776 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 4 |
Hb_000620_020 |
0.1089976237 |
- |
- |
PREDICTED: uncharacterized protein LOC105649917 [Jatropha curcas] |
| 5 |
Hb_001155_050 |
0.109073065 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 6 |
Hb_001511_030 |
0.1107615985 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 [Jatropha curcas] |
| 7 |
Hb_002871_040 |
0.1140165309 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 8 |
Hb_006438_020 |
0.1143287521 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 9 |
Hb_061256_010 |
0.1187031506 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000086_270 |
0.1220392553 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 11 |
Hb_000818_120 |
0.1241624539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000526_010 |
0.1243885635 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 13 |
Hb_000589_170 |
0.1291832505 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 14 |
Hb_006588_190 |
0.1298947614 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 15 |
Hb_000235_070 |
0.1301750663 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 16 |
Hb_017193_010 |
0.1315308293 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001160_110 |
0.1327665127 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 18 |
Hb_011381_040 |
0.1336912821 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 19 |
Hb_013405_070 |
0.1374267472 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 20 |
Hb_001102_070 |
0.1385695851 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |