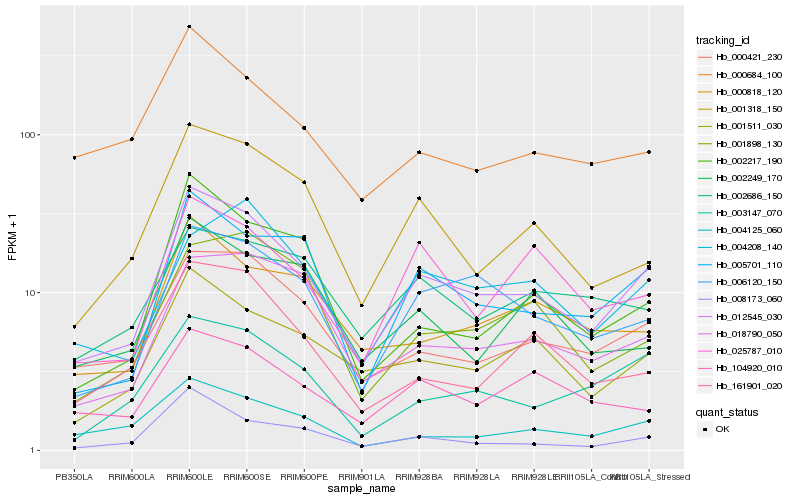
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012545_030 |
0.0 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 2 |
Hb_000421_230 |
0.12023957 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 3 |
Hb_005701_110 |
0.1211231666 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 4 |
Hb_006120_150 |
0.1221477463 |
- |
- |
invertase [Hevea brasiliensis] |
| 5 |
Hb_001511_030 |
0.1260426836 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 [Jatropha curcas] |
| 6 |
Hb_001318_150 |
0.1317808707 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 7 |
Hb_003147_070 |
0.1404679814 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 8 |
Hb_018790_050 |
0.1486109019 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 9 |
Hb_004125_060 |
0.1497628407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000818_120 |
0.1502653864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_025787_010 |
0.1504230147 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 12 |
Hb_104920_010 |
0.1520840347 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 13 |
Hb_161901_020 |
0.153594764 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 14 |
Hb_000684_100 |
0.1564549604 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 15 |
Hb_008173_060 |
0.1566721273 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 16 |
Hb_002217_190 |
0.1566923578 |
- |
- |
invertase [Hevea brasiliensis] |
| 17 |
Hb_004208_140 |
0.1580748565 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 18 |
Hb_001898_130 |
0.1612907103 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_002249_170 |
0.1617706812 |
- |
- |
copine, putative [Ricinus communis] |
| 20 |
Hb_002686_150 |
0.1629581577 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |