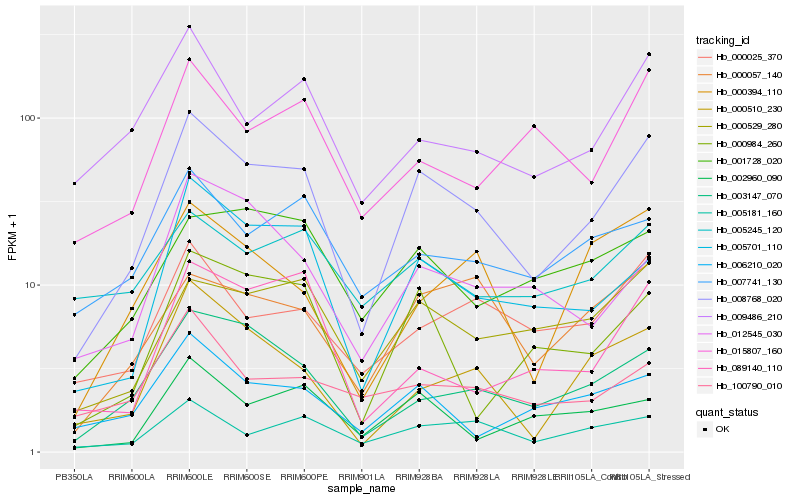
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008768_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 2 |
Hb_005701_110 |
0.1498307092 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 3 |
Hb_003147_070 |
0.1596690907 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 4 |
Hb_000529_280 |
0.1624156801 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005181_160 |
0.164139287 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 6 |
Hb_000057_140 |
0.1687339159 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 7 |
Hb_000025_370 |
0.1690418335 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009486_210 |
0.1710583952 |
- |
- |
- |
| 9 |
Hb_000510_230 |
0.174475405 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 10 |
Hb_000984_260 |
0.1760181216 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 11 |
Hb_006210_020 |
0.1806283331 |
- |
- |
PREDICTED: calcium sensing receptor, chloroplastic [Jatropha curcas] |
| 12 |
Hb_007741_130 |
0.1842589722 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 13 |
Hb_002960_090 |
0.1858649304 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000394_110 |
0.1861326062 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001728_020 |
0.1867742097 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 16 |
Hb_015807_160 |
0.1879140606 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 17 |
Hb_012545_030 |
0.19026528 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 18 |
Hb_089140_110 |
0.1943944385 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 19 |
Hb_100790_010 |
0.1974935167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005245_120 |
0.1998583152 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |