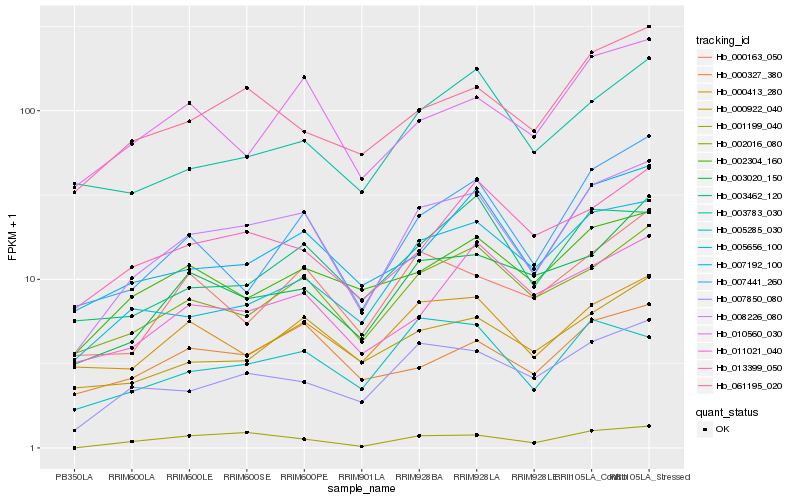
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008226_080 |
0.0 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 2 |
Hb_002016_080 |
0.1097098115 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007850_080 |
0.1098374744 |
- |
- |
PREDICTED: auxilin-like protein 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007192_100 |
0.1215436364 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 5 |
Hb_000413_280 |
0.1245038342 |
- |
- |
PREDICTED: probable UDP-3-O-acylglucosamine N-acyltransferase 2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_061195_020 |
0.1251883151 |
- |
- |
PREDICTED: uncharacterized protein LOC101990594 [Microtus ochrogaster] |
| 7 |
Hb_000922_040 |
0.1262269602 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 8 |
Hb_001199_040 |
0.1283327537 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_003020_150 |
0.1293288063 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 10 |
Hb_002304_160 |
0.1293633942 |
- |
- |
PREDICTED: nudix hydrolase 15, mitochondrial-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_007441_260 |
0.1313586504 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000163_050 |
0.131875659 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 13 |
Hb_010560_030 |
0.1321907269 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |
| 14 |
Hb_003462_120 |
0.1349524086 |
- |
- |
Acidic endochitinase [Theobroma cacao] |
| 15 |
Hb_000327_380 |
0.1350960694 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 16 |
Hb_005285_030 |
0.1366347457 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 17 |
Hb_011021_040 |
0.1368951699 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 18 |
Hb_005656_100 |
0.1372414559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_013399_050 |
0.1377293198 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_003783_030 |
0.1386755408 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |