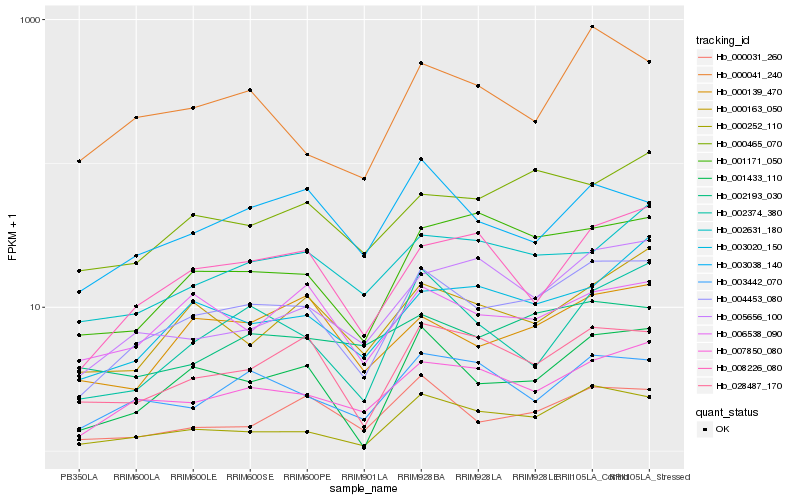
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004453_080 |
0.0 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 2 |
Hb_007850_080 |
0.1103155773 |
- |
- |
PREDICTED: auxilin-like protein 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001433_110 |
0.1211654176 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000252_110 |
0.127714181 |
- |
- |
- |
| 5 |
Hb_028487_170 |
0.1332599455 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 6 |
Hb_003442_070 |
0.1369385106 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 7 |
Hb_008226_080 |
0.1407513852 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 8 |
Hb_000139_470 |
0.1459480255 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_000465_070 |
0.1459737111 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase small subunit 3-like [Jatropha curcas] |
| 10 |
Hb_000163_050 |
0.1468105293 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 11 |
Hb_000031_260 |
0.1483746178 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 12 |
Hb_002631_180 |
0.1486486194 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 13 |
Hb_001171_050 |
0.1493061893 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |
| 14 |
Hb_000041_240 |
0.1512318119 |
- |
- |
glutaredoxin [Hevea brasiliensis] |
| 15 |
Hb_006538_090 |
0.1520793068 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 16 |
Hb_003020_150 |
0.152124315 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 17 |
Hb_002374_380 |
0.1524655362 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_005656_100 |
0.1525750543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002193_030 |
0.1557293827 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 20 |
Hb_003038_140 |
0.1560442587 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |