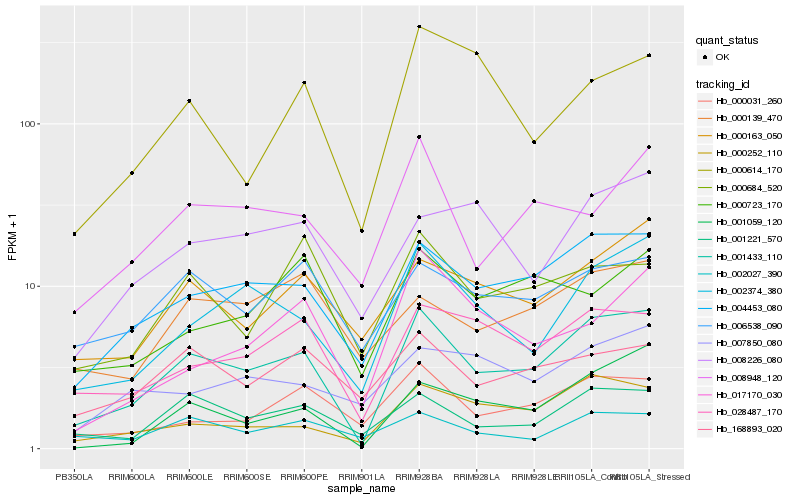
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001433_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004453_080 |
0.1211654176 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 3 |
Hb_002374_380 |
0.1477237999 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_001221_570 |
0.1550257238 |
- |
- |
- |
| 5 |
Hb_000252_110 |
0.156827261 |
- |
- |
- |
| 6 |
Hb_028487_170 |
0.1581934787 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 7 |
Hb_001059_120 |
0.1654142368 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000031_260 |
0.1666592293 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 9 |
Hb_000163_050 |
0.169901081 |
- |
- |
SPLICEOSOMAL protein U1A [Populus trichocarpa] |
| 10 |
Hb_008948_120 |
0.1793741556 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_006538_090 |
0.1836404286 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 12 |
Hb_008226_080 |
0.1850045573 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 13 |
Hb_168893_020 |
0.1853011022 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 14 |
Hb_002027_390 |
0.1881645564 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000684_520 |
0.1887400138 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 16 |
Hb_000139_470 |
0.1894655536 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_000614_170 |
0.1919341549 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 18 |
Hb_017170_030 |
0.1930738156 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 19 |
Hb_000723_170 |
0.1938327414 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 20 |
Hb_007850_080 |
0.1947403835 |
- |
- |
PREDICTED: auxilin-like protein 1 isoform X1 [Jatropha curcas] |