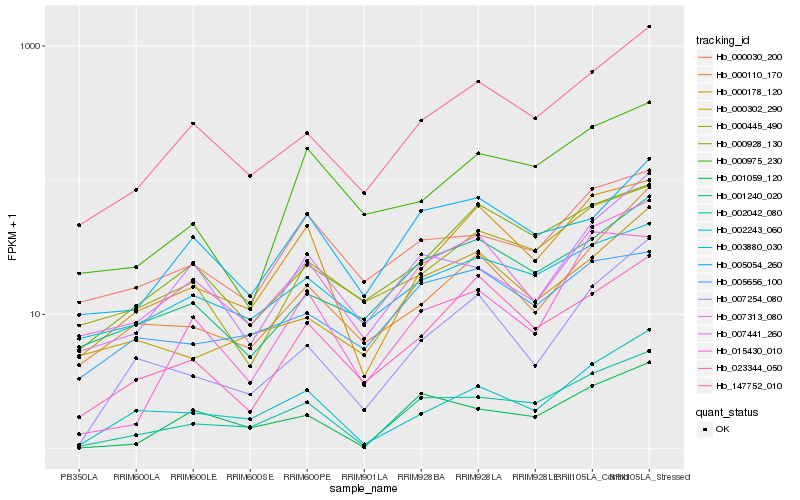
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002042_080 |
0.0 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 2 |
Hb_001059_120 |
0.1196205503 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000178_120 |
0.1349739921 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 4 |
Hb_147752_010 |
0.1357936833 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 5 |
Hb_002243_060 |
0.1387413375 |
- |
- |
hypothetical protein POPTR_0022s00750g [Populus trichocarpa] |
| 6 |
Hb_007441_260 |
0.1624669708 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_001240_020 |
0.1656546144 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 8 |
Hb_015430_010 |
0.167380476 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 9 |
Hb_000302_290 |
0.167903332 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 10 |
Hb_000030_200 |
0.1704649677 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 11 |
Hb_007313_080 |
0.1710357358 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 12 |
Hb_007254_080 |
0.1728894213 |
- |
- |
- |
| 13 |
Hb_023344_050 |
0.1750983446 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 14 |
Hb_005054_260 |
0.1756117155 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 15 |
Hb_000445_490 |
0.1759183988 |
- |
- |
- |
| 16 |
Hb_000928_130 |
0.1785632835 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 17 |
Hb_000110_170 |
0.1804907845 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_005656_100 |
0.1806356984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003880_030 |
0.1829731986 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000975_230 |
0.1833977398 |
- |
- |
hypothetical protein ZEAMMB73_864543 [Zea mays] |