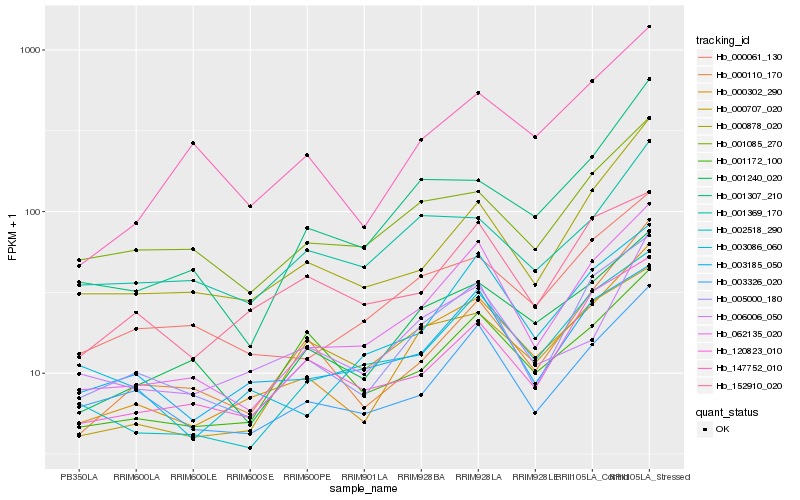
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_290 |
0.0 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 2 |
Hb_001240_020 |
0.0942615624 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 3 |
Hb_005000_180 |
0.0947197298 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 4 |
Hb_062135_020 |
0.1031580628 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Nicotiana sylvestris] |
| 5 |
Hb_000061_130 |
0.1134834507 |
- |
- |
rotamase, putative [Ricinus communis] |
| 6 |
Hb_147752_010 |
0.1218600998 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 7 |
Hb_002518_290 |
0.1232586885 |
- |
- |
Acyl-coenzyme A thioesterase 13 [Gossypium arboreum] |
| 8 |
Hb_000110_170 |
0.124220037 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_001307_210 |
0.1280327641 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 10 |
Hb_006006_050 |
0.1289083257 |
- |
- |
PREDICTED: methylthioribose kinase-like [Gossypium raimondii] |
| 11 |
Hb_120823_010 |
0.129014647 |
- |
- |
- |
| 12 |
Hb_001085_270 |
0.1296937058 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 13 |
Hb_000878_020 |
0.1299305421 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 14 |
Hb_000707_020 |
0.131769521 |
- |
- |
hypothetical protein B456_002G255500 [Gossypium raimondii] |
| 15 |
Hb_003185_050 |
0.1342787825 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 16 |
Hb_003086_060 |
0.1345177256 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 17 |
Hb_003326_020 |
0.134634001 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 18 |
Hb_001369_170 |
0.1366021248 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1-like [Gossypium raimondii] |
| 19 |
Hb_001172_100 |
0.1376574816 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_152910_020 |
0.1381905205 |
- |
- |
PREDICTED: lactoylglutathione lyase-like [Jatropha curcas] |