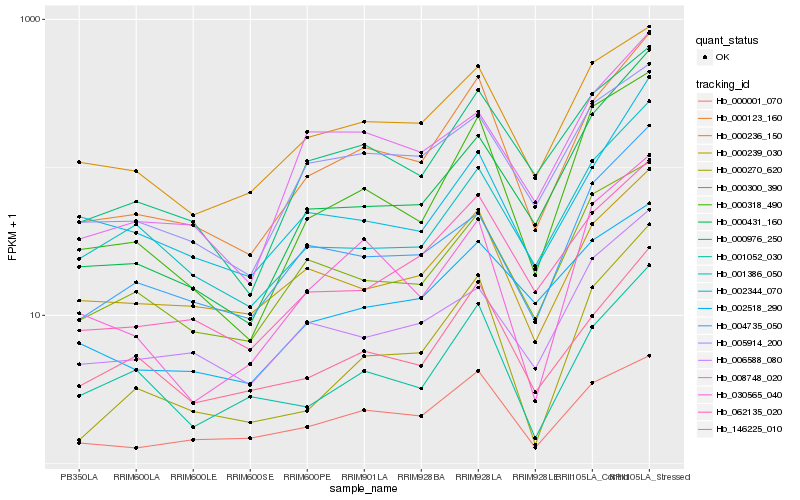
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000123_160 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 2 |
Hb_062135_020 |
0.0948631417 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Nicotiana sylvestris] |
| 3 |
Hb_000976_250 |
0.0993103058 |
- |
- |
- |
| 4 |
Hb_146225_010 |
0.1014268548 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 5 |
Hb_000318_490 |
0.1036479556 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 6 |
Hb_000236_150 |
0.1141538273 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 7 |
Hb_000001_070 |
0.118699285 |
- |
- |
- |
| 8 |
Hb_005914_200 |
0.1191297191 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 9 |
Hb_001052_030 |
0.1228379361 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000239_030 |
0.1230060759 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 11 |
Hb_001386_050 |
0.1253259505 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 12 |
Hb_002344_070 |
0.1267526638 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 13 |
Hb_000300_390 |
0.1270892148 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 14 |
Hb_000270_620 |
0.1280040845 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004735_050 |
0.1282763231 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 16 |
Hb_030565_040 |
0.1285436749 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_008748_020 |
0.1287641892 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 18 |
Hb_002518_290 |
0.1316587862 |
- |
- |
Acyl-coenzyme A thioesterase 13 [Gossypium arboreum] |
| 19 |
Hb_000431_160 |
0.1316860511 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 20 |
Hb_006588_080 |
0.1349081854 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |