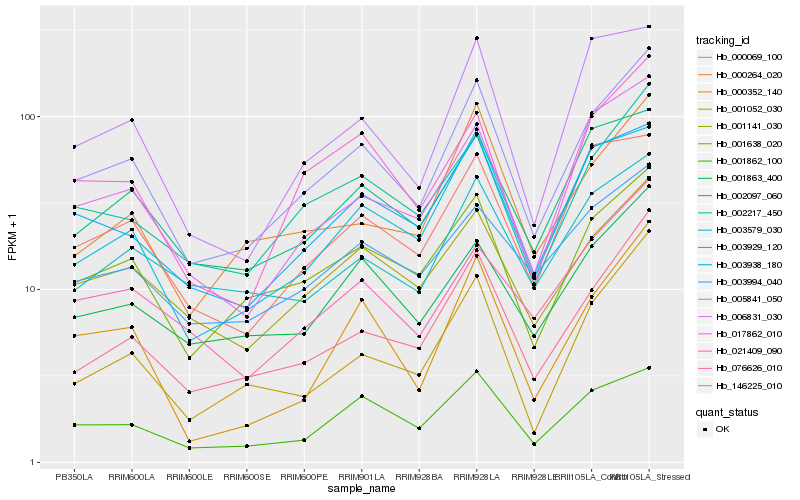
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005841_050 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP1-like [Jatropha curcas] |
| 2 |
Hb_002217_450 |
0.0835120433 |
- |
- |
PREDICTED: probable prefoldin subunit 3 [Jatropha curcas] |
| 3 |
Hb_146225_010 |
0.0843834525 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 4 |
Hb_001638_020 |
0.0844348237 |
- |
- |
PREDICTED: protein OSB1, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_001141_030 |
0.0898837808 |
- |
- |
hypothetical protein PRUPE_ppa024854mg [Prunus persica] |
| 6 |
Hb_017862_010 |
0.095933304 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 7 |
Hb_021409_090 |
0.0964716891 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0003s14450g [Populus trichocarpa] |
| 8 |
Hb_001863_400 |
0.0989952523 |
- |
- |
hypothetical protein EUGRSUZ_F03302 [Eucalyptus grandis] |
| 9 |
Hb_003938_180 |
0.1021604758 |
- |
- |
lesion inducing family protein [Populus trichocarpa] |
| 10 |
Hb_001862_100 |
0.1031377981 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003929_120 |
0.1062112338 |
- |
- |
PREDICTED: 50S ribosomal protein L3-2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002097_060 |
0.1064521595 |
- |
- |
PREDICTED: 50S ribosomal protein L9, chloroplastic-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000264_020 |
0.1072552437 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_076626_010 |
0.1084027446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000352_140 |
0.1088897487 |
- |
- |
- |
| 16 |
Hb_000069_100 |
0.1093214192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001052_030 |
0.1093875141 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003994_040 |
0.1108251016 |
- |
- |
- |
| 19 |
Hb_003579_030 |
0.1119221927 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 20 |
Hb_006831_030 |
0.1123806139 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |