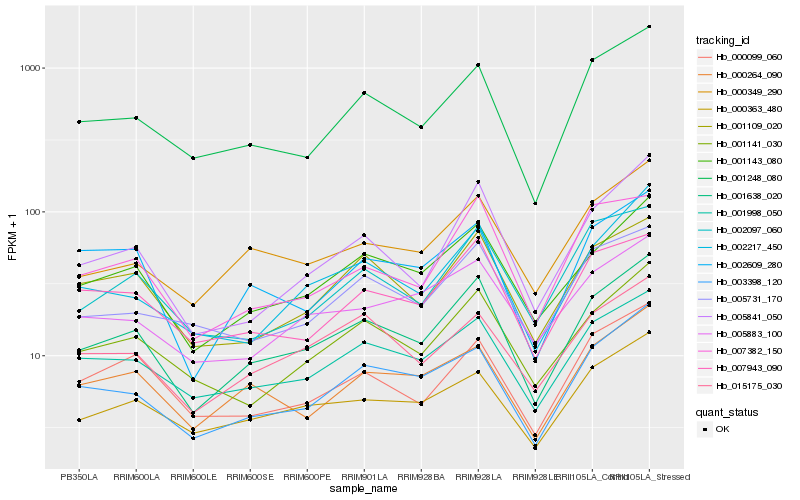
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_020 |
0.0 |
- |
- |
PREDICTED: protein OSB1, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_001143_080 |
0.065515018 |
- |
- |
PREDICTED: F-box protein 7 [Jatropha curcas] |
| 3 |
Hb_003398_120 |
0.0810728101 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 4 |
Hb_001998_050 |
0.0829241961 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_005841_050 |
0.0844348237 |
- |
- |
PREDICTED: F-box protein SKIP1-like [Jatropha curcas] |
| 6 |
Hb_001248_080 |
0.0844936208 |
- |
- |
PREDICTED: 40S ribosomal protein S29 [Nelumbo nucifera] |
| 7 |
Hb_001141_030 |
0.0863133963 |
- |
- |
hypothetical protein PRUPE_ppa024854mg [Prunus persica] |
| 8 |
Hb_001109_020 |
0.0880507345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000349_290 |
0.0882916946 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |
| 10 |
Hb_002217_450 |
0.0900050415 |
- |
- |
PREDICTED: probable prefoldin subunit 3 [Jatropha curcas] |
| 11 |
Hb_000363_480 |
0.0919822669 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_007382_150 |
0.0962288276 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |
| 13 |
Hb_000264_090 |
0.0963329282 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 14 |
Hb_015175_030 |
0.0980023177 |
- |
- |
PREDICTED: RNA-binding protein 24-A [Jatropha curcas] |
| 15 |
Hb_000099_060 |
0.0984296043 |
- |
- |
hypothetical protein JCGZ_23948 [Jatropha curcas] |
| 16 |
Hb_007943_090 |
0.0990112834 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 17 |
Hb_002609_280 |
0.0998105061 |
- |
- |
villin 1-4, putative [Ricinus communis] |
| 18 |
Hb_002097_060 |
0.1000566223 |
- |
- |
PREDICTED: 50S ribosomal protein L9, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005731_170 |
0.1006468478 |
- |
- |
hypothetical protein PRUPE_ppa011750mg [Prunus persica] |
| 20 |
Hb_005883_100 |
0.1019526483 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |