| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
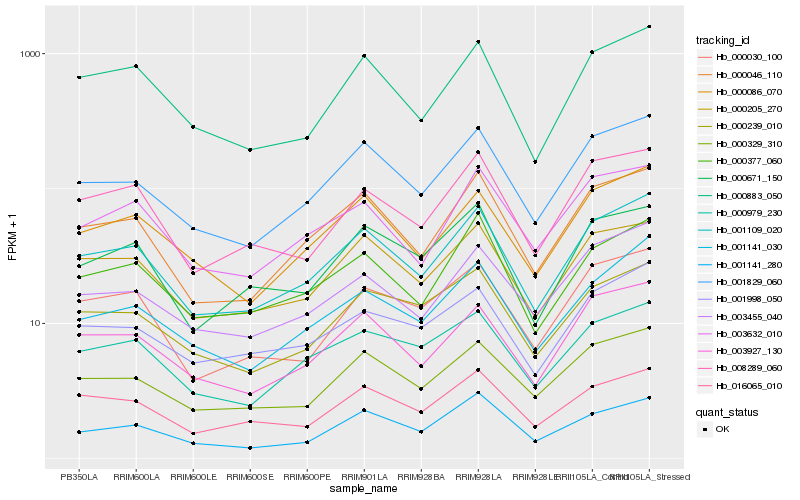
Hb_001109_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001829_060 |
0.0489862685 |
- |
- |
Eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 3 |
Hb_000046_110 |
0.0502126166 |
- |
- |
PREDICTED: 54S ribosomal protein L39, mitochondrial [Jatropha curcas] |
| 4 |
Hb_003927_130 |
0.0600757759 |
- |
- |
PREDICTED: nudix hydrolase 9 [Phoenix dactylifera] |
| 5 |
Hb_000205_270 |
0.0627533775 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000883_050 |
0.0630307863 |
- |
- |
PREDICTED: 60S ribosomal protein L37a-like, partial [Solanum tuberosum] |
| 7 |
Hb_000086_070 |
0.0646757419 |
- |
- |
Peptidase S24/S26A/S26B/S26C family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_000329_310 |
0.0652551559 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000377_060 |
0.0655421926 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 10 |
Hb_001141_030 |
0.0692613782 |
- |
- |
hypothetical protein PRUPE_ppa024854mg [Prunus persica] |
| 11 |
Hb_008289_060 |
0.0709267998 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_016065_010 |
0.0734251387 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08305 [Jatropha curcas] |
| 13 |
Hb_000979_230 |
0.0737142796 |
- |
- |
PREDICTED: protein BCCIP homolog [Jatropha curcas] |
| 14 |
Hb_001998_050 |
0.0741056353 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_003455_040 |
0.0751372561 |
- |
- |
hypothetical protein JCGZ_17270 [Jatropha curcas] |
| 16 |
Hb_001141_280 |
0.0764531433 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Vitis vinifera] |
| 17 |
Hb_000030_100 |
0.0768871404 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |
| 18 |
Hb_000671_150 |
0.077136181 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 19 |
Hb_000239_010 |
0.0774222623 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_003632_010 |
0.0777187603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |