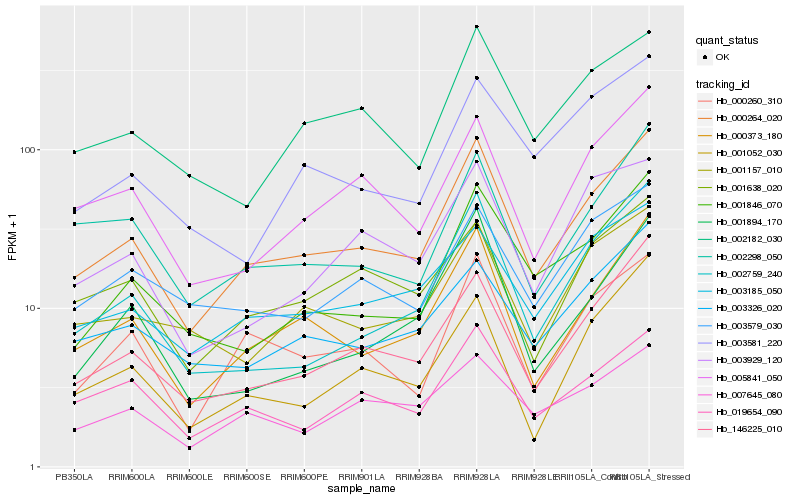
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_020 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000373_180 |
0.0766855692 |
transcription factor |
TF Family: Trihelix |
hypothetical protein JCGZ_25182 [Jatropha curcas] |
| 3 |
Hb_146225_010 |
0.0854739102 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 4 |
Hb_000260_310 |
0.0991943041 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 5 |
Hb_005841_050 |
0.1072552437 |
- |
- |
PREDICTED: F-box protein SKIP1-like [Jatropha curcas] |
| 6 |
Hb_001846_070 |
0.1081581609 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 7 |
Hb_002759_240 |
0.1116404157 |
- |
- |
hypothetical protein EUGRSUZ_E01319 [Eucalyptus grandis] |
| 8 |
Hb_019654_090 |
0.1116962025 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33680 [Jatropha curcas] |
| 9 |
Hb_003185_050 |
0.1117224301 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 10 |
Hb_001157_010 |
0.1130711968 |
- |
- |
deoxyhypusine synthase, putative [Ricinus communis] |
| 11 |
Hb_002298_050 |
0.1133019801 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 12 |
Hb_003326_020 |
0.1146171085 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 13 |
Hb_002182_030 |
0.1146656261 |
- |
- |
hypothetical protein Csa_5G139175 [Cucumis sativus] |
| 14 |
Hb_003579_030 |
0.1214388545 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 15 |
Hb_007645_080 |
0.1218604615 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 16 |
Hb_001638_020 |
0.1221007019 |
- |
- |
PREDICTED: protein OSB1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_001052_030 |
0.1234496303 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003929_120 |
0.1241451443 |
- |
- |
PREDICTED: 50S ribosomal protein L3-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001894_170 |
0.1253316997 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |
| 20 |
Hb_003581_220 |
0.1297082654 |
- |
- |
unnamed protein product [Vitis vinifera] |