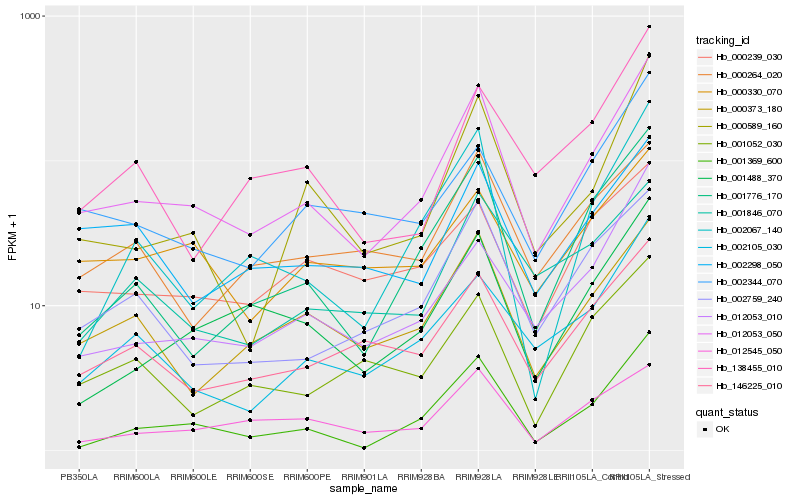
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012053_050 |
0.0 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 2 |
Hb_001369_600 |
0.1206114848 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_001776_170 |
0.1270625868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000589_160 |
0.1310484853 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 5 |
Hb_000373_180 |
0.1316574972 |
transcription factor |
TF Family: Trihelix |
hypothetical protein JCGZ_25182 [Jatropha curcas] |
| 6 |
Hb_002759_240 |
0.1345957087 |
- |
- |
hypothetical protein EUGRSUZ_E01319 [Eucalyptus grandis] |
| 7 |
Hb_001488_370 |
0.1357168549 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 8 |
Hb_146225_010 |
0.1381805418 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 9 |
Hb_002105_030 |
0.1424522881 |
- |
- |
- |
| 10 |
Hb_002067_140 |
0.1461967974 |
- |
- |
- |
| 11 |
Hb_012053_010 |
0.1473220369 |
- |
- |
srpk, putative [Ricinus communis] |
| 12 |
Hb_002344_070 |
0.1529238807 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 13 |
Hb_138455_010 |
0.1533752893 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_002298_050 |
0.1535554165 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001052_030 |
0.1554811175 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001846_070 |
0.155767681 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 17 |
Hb_000239_030 |
0.1561605621 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 18 |
Hb_012545_050 |
0.1565482536 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Jatropha curcas] |
| 19 |
Hb_000264_020 |
0.1571604694 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000330_070 |
0.1588767657 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |