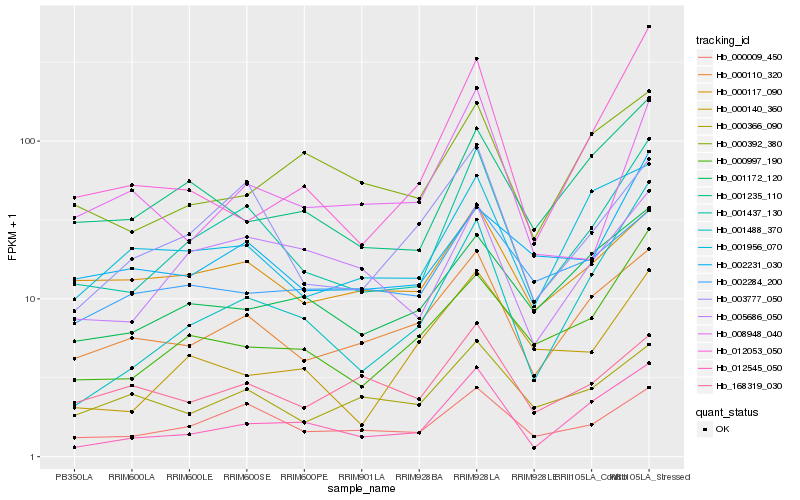
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_130 |
0.0 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 2 |
Hb_000009_450 |
0.130167139 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_012545_050 |
0.1336594116 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Jatropha curcas] |
| 4 |
Hb_003777_050 |
0.1370497512 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 5 |
Hb_000110_320 |
0.138367367 |
- |
- |
PREDICTED: ceramide-1-phosphate transfer protein [Jatropha curcas] |
| 6 |
Hb_001488_370 |
0.1467155205 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 7 |
Hb_001235_110 |
0.1540947508 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 8 |
Hb_005686_050 |
0.1551036656 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 9 |
Hb_000997_190 |
0.155247524 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like [Jatropha curcas] |
| 10 |
Hb_001956_070 |
0.16113386 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000117_090 |
0.1652597235 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 12 |
Hb_001172_120 |
0.1659822213 |
- |
- |
hypothetical protein B456_012G082000 [Gossypium raimondii] |
| 13 |
Hb_002231_030 |
0.1705887859 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 14 |
Hb_000366_090 |
0.1754541872 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |
| 15 |
Hb_008948_040 |
0.1822521854 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 16 |
Hb_002284_200 |
0.1826159139 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 17 |
Hb_168319_030 |
0.1832255808 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 18 |
Hb_000392_380 |
0.1836727205 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 19 |
Hb_012053_050 |
0.1859175893 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 20 |
Hb_000140_360 |
0.1861921932 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |