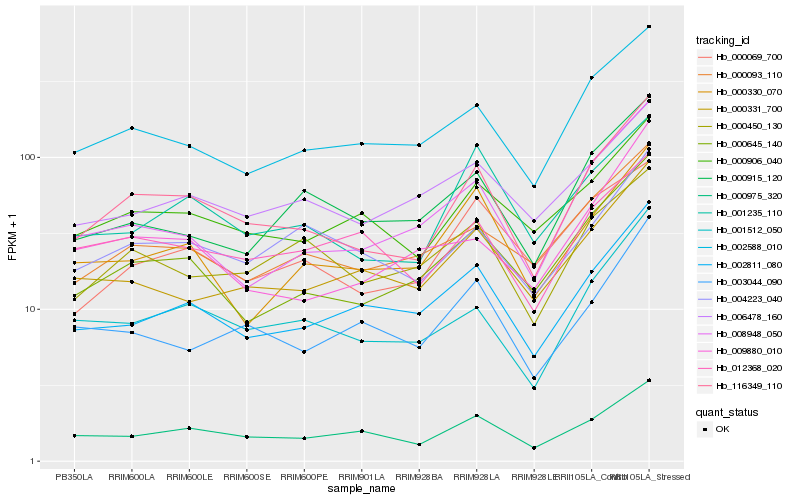
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116349_110 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 2 |
Hb_000645_140 |
0.0950923376 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A [Nelumbo nucifera] |
| 3 |
Hb_001512_050 |
0.1005753485 |
- |
- |
copine, putative [Ricinus communis] |
| 4 |
Hb_002588_010 |
0.1049580305 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 5 |
Hb_000975_320 |
0.1089495201 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002811_080 |
0.1119764705 |
- |
- |
hypothetical protein POPTR_0001s08090g [Populus trichocarpa] |
| 7 |
Hb_000906_040 |
0.1132803642 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001235_110 |
0.1144011307 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 9 |
Hb_000093_110 |
0.1163055256 |
- |
- |
PREDICTED: cytochrome b5 isoform A [Jatropha curcas] |
| 10 |
Hb_008948_050 |
0.1173724749 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |
| 11 |
Hb_000331_700 |
0.124532104 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 12 |
Hb_000330_070 |
0.1246269468 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 13 |
Hb_003044_090 |
0.1269114772 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 14 |
Hb_000069_700 |
0.1279001502 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 15 |
Hb_004223_040 |
0.1293172309 |
- |
- |
PREDICTED: AP-2 complex subunit sigma [Fragaria vesca subsp. vesca] |
| 16 |
Hb_012368_020 |
0.1294663568 |
- |
- |
PREDICTED: uncharacterized protein LOC105650964 [Jatropha curcas] |
| 17 |
Hb_006478_160 |
0.1309891252 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 18 |
Hb_000915_120 |
0.1315205336 |
- |
- |
Peptidylprolyl cis/trans isomerase [Theobroma cacao] |
| 19 |
Hb_000450_130 |
0.1316822112 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 20 |
Hb_009880_010 |
0.1350200707 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |