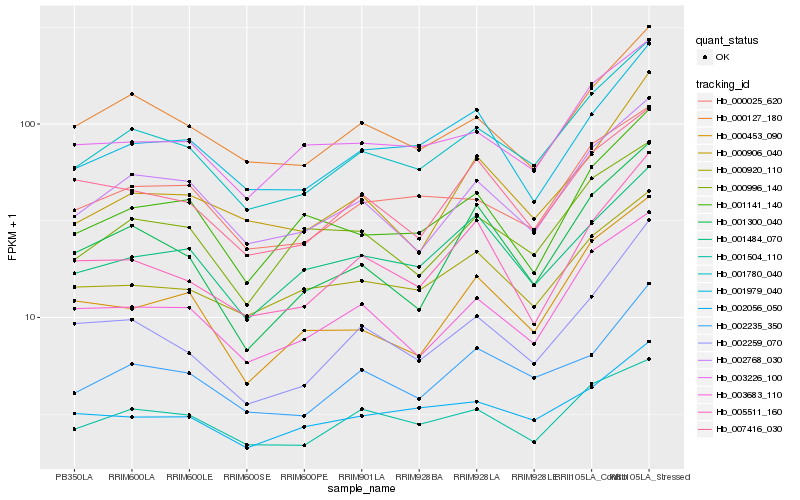
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001780_040 |
0.0 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 2 |
Hb_002768_030 |
0.0568812632 |
- |
- |
gaba(A) receptor-associated protein, putative [Ricinus communis] |
| 3 |
Hb_003683_110 |
0.0643427657 |
- |
- |
JHL10I11.3 [Jatropha curcas] |
| 4 |
Hb_002235_350 |
0.0645674849 |
- |
- |
PREDICTED: uncharacterized protein LOC105635929 [Jatropha curcas] |
| 5 |
Hb_000025_620 |
0.0655524 |
- |
- |
hypothetical protein POPTR_0009s05200g [Populus trichocarpa] |
| 6 |
Hb_000127_180 |
0.0671522925 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 7 |
Hb_003226_100 |
0.0717535495 |
- |
- |
PREDICTED: 60S ribosomal protein L18-2 [Jatropha curcas] |
| 8 |
Hb_001979_040 |
0.0782392775 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1-like [Gossypium raimondii] |
| 9 |
Hb_002259_070 |
0.0783367282 |
- |
- |
taz protein, putative [Ricinus communis] |
| 10 |
Hb_000996_140 |
0.0802039012 |
- |
- |
PREDICTED: EKC/KEOPS complex subunit Tprkb [Jatropha curcas] |
| 11 |
Hb_000920_110 |
0.0813033572 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP4 [Jatropha curcas] |
| 12 |
Hb_001504_110 |
0.0823997671 |
- |
- |
PREDICTED: uncharacterized protein LOC105630336 [Jatropha curcas] |
| 13 |
Hb_001141_140 |
0.0832851839 |
- |
- |
20S proteasome beta subunit D3 [Hevea brasiliensis] |
| 14 |
Hb_001300_040 |
0.0834008988 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 15 |
Hb_001484_070 |
0.0848149602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000906_040 |
0.084853015 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007416_030 |
0.0851254719 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 10 [Jatropha curcas] |
| 18 |
Hb_005511_160 |
0.0858398403 |
- |
- |
OB-fold nucleic acid binding domain-containing protein [Theobroma cacao] |
| 19 |
Hb_000453_090 |
0.0859179709 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 20 |
Hb_002056_050 |
0.0864397778 |
- |
- |
PREDICTED: uncharacterized protein LOC105130744 [Populus euphratica] |