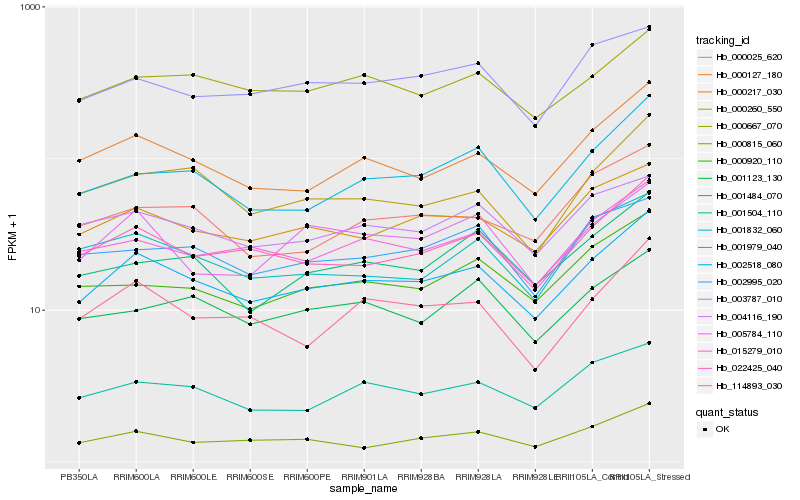
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002518_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639837 [Jatropha curcas] |
| 2 |
Hb_022425_040 |
0.0596896003 |
- |
- |
PREDICTED: probable RNA methyltransferase At5g51130 [Jatropha curcas] |
| 3 |
Hb_000127_180 |
0.0672490044 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 4 |
Hb_000217_030 |
0.0718312459 |
- |
- |
Leukocyte immunoglobulin-like receptor subfamily A member 5 isoform 1 [Theobroma cacao] |
| 5 |
Hb_015279_010 |
0.0736531827 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 6 |
Hb_114893_030 |
0.0797736117 |
- |
- |
PREDICTED: ribosome-recycling factor, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001504_110 |
0.0798188681 |
- |
- |
PREDICTED: uncharacterized protein LOC105630336 [Jatropha curcas] |
| 8 |
Hb_000260_550 |
0.0801503765 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 9 |
Hb_001979_040 |
0.0805751887 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1-like [Gossypium raimondii] |
| 10 |
Hb_000815_060 |
0.0811516444 |
- |
- |
PREDICTED: uncharacterized protein LOC105630460 [Jatropha curcas] |
| 11 |
Hb_000667_070 |
0.0826572197 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 12 |
Hb_004116_190 |
0.0860791492 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 2 [Jatropha curcas] |
| 13 |
Hb_003787_010 |
0.0870299039 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Phoenix dactylifera] |
| 14 |
Hb_000920_110 |
0.0870546442 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP4 [Jatropha curcas] |
| 15 |
Hb_001832_060 |
0.0878359432 |
- |
- |
PREDICTED: translation machinery-associated protein 22 [Jatropha curcas] |
| 16 |
Hb_001484_070 |
0.08978298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001123_130 |
0.0901316274 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_005784_110 |
0.0905521267 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_002995_020 |
0.0919661573 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 2 [Jatropha curcas] |
| 20 |
Hb_000025_620 |
0.0928560769 |
- |
- |
hypothetical protein POPTR_0009s05200g [Populus trichocarpa] |