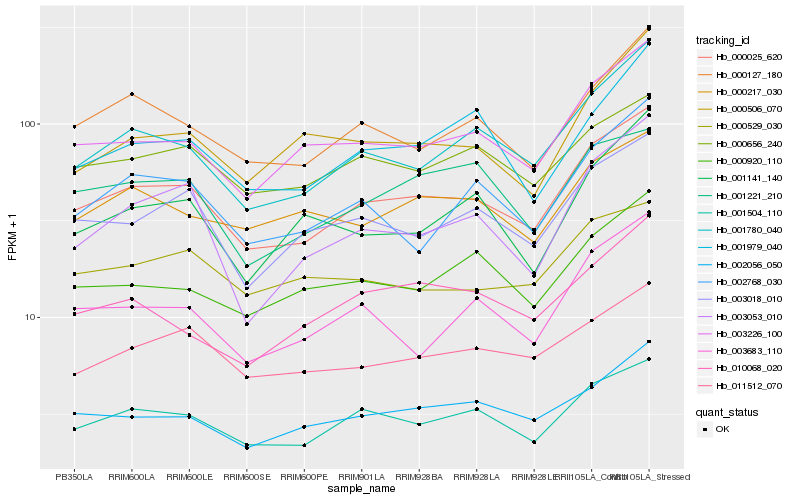
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_620 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s05200g [Populus trichocarpa] |
| 2 |
Hb_001504_110 |
0.0546022936 |
- |
- |
PREDICTED: uncharacterized protein LOC105630336 [Jatropha curcas] |
| 3 |
Hb_003226_100 |
0.063695943 |
- |
- |
PREDICTED: 60S ribosomal protein L18-2 [Jatropha curcas] |
| 4 |
Hb_001780_040 |
0.0655524 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 5 |
Hb_003018_010 |
0.0696756137 |
- |
- |
PREDICTED: 6,7-dimethyl-8-ribityllumazine synthase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000127_180 |
0.0706870183 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 7 |
Hb_002056_050 |
0.0732978828 |
- |
- |
PREDICTED: uncharacterized protein LOC105130744 [Populus euphratica] |
| 8 |
Hb_011512_070 |
0.0791930337 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 9 |
Hb_002768_030 |
0.0824133435 |
- |
- |
gaba(A) receptor-associated protein, putative [Ricinus communis] |
| 10 |
Hb_000920_110 |
0.0826823278 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP4 [Jatropha curcas] |
| 11 |
Hb_000656_240 |
0.0834975712 |
- |
- |
proteasome subunit alpha type, putative [Ricinus communis] |
| 12 |
Hb_003683_110 |
0.0839594343 |
- |
- |
JHL10I11.3 [Jatropha curcas] |
| 13 |
Hb_001979_040 |
0.085034792 |
- |
- |
PREDICTED: 60S ribosomal protein L21-1-like [Gossypium raimondii] |
| 14 |
Hb_000217_030 |
0.0856515069 |
- |
- |
Leukocyte immunoglobulin-like receptor subfamily A member 5 isoform 1 [Theobroma cacao] |
| 15 |
Hb_003053_010 |
0.0856749264 |
- |
- |
PREDICTED: uncharacterized protein LOC105641481 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000529_030 |
0.0893054951 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_010068_020 |
0.0899786032 |
- |
- |
PREDICTED: uncharacterized protein LOC105632966 [Jatropha curcas] |
| 18 |
Hb_001221_210 |
0.0902063139 |
- |
- |
thiF family protein [Populus trichocarpa] |
| 19 |
Hb_000506_070 |
0.0915422944 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 20 |
Hb_001141_140 |
0.0919895836 |
- |
- |
20S proteasome beta subunit D3 [Hevea brasiliensis] |