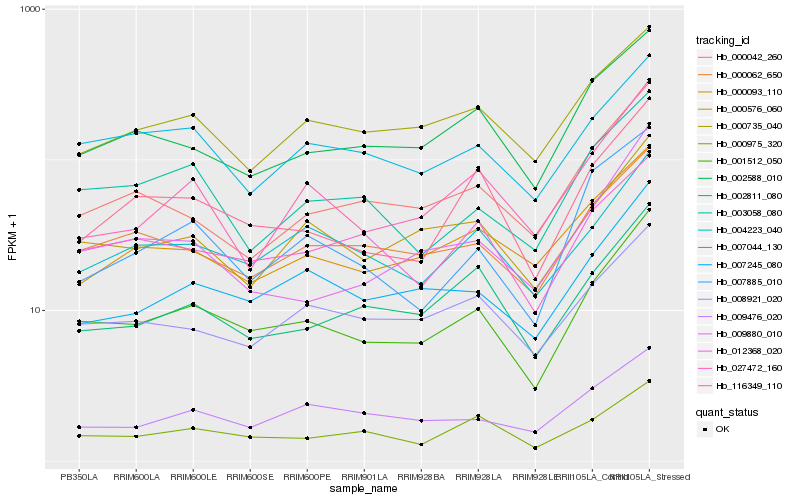
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001512_050 |
0.0 |
- |
- |
copine, putative [Ricinus communis] |
| 2 |
Hb_007245_080 |
0.0976701996 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 3 |
Hb_116349_110 |
0.1005753485 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 4 |
Hb_000062_650 |
0.1015077075 |
- |
- |
PREDICTED: uncharacterized protein LOC105628245 [Jatropha curcas] |
| 5 |
Hb_000576_060 |
0.103994564 |
- |
- |
truncated hemoglobin [Populus tremula x Populus tremuloides] |
| 6 |
Hb_002588_010 |
0.1053563751 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 7 |
Hb_004223_040 |
0.1057945535 |
- |
- |
PREDICTED: AP-2 complex subunit sigma [Fragaria vesca subsp. vesca] |
| 8 |
Hb_000975_320 |
0.1097699749 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007044_130 |
0.1120639768 |
- |
- |
PREDICTED: cyclin-B1-2 [Jatropha curcas] |
| 10 |
Hb_003058_080 |
0.1128472527 |
- |
- |
Chaperonin 10 isoform 1 [Theobroma cacao] |
| 11 |
Hb_002811_080 |
0.1129797819 |
- |
- |
hypothetical protein POPTR_0001s08090g [Populus trichocarpa] |
| 12 |
Hb_009880_010 |
0.1134727332 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 13 |
Hb_000735_040 |
0.113632022 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 14 |
Hb_008921_020 |
0.1155226769 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 1 homolog [Jatropha curcas] |
| 15 |
Hb_012368_020 |
0.1163814556 |
- |
- |
PREDICTED: uncharacterized protein LOC105650964 [Jatropha curcas] |
| 16 |
Hb_000042_260 |
0.1168942975 |
- |
- |
PREDICTED: uncharacterized protein LOC105632814 [Jatropha curcas] |
| 17 |
Hb_000093_110 |
0.1174018285 |
- |
- |
PREDICTED: cytochrome b5 isoform A [Jatropha curcas] |
| 18 |
Hb_009476_020 |
0.1192848988 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 19 |
Hb_027472_160 |
0.1198816151 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 20 |
Hb_007885_010 |
0.1237651305 |
- |
- |
60S ribosomal protein L24-1 isoform 1 [Theobroma cacao] |