| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
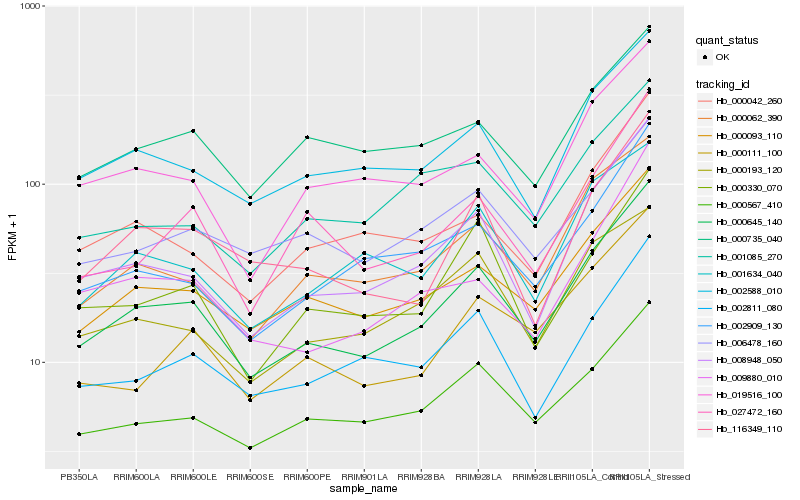
Hb_000645_140 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A [Nelumbo nucifera] |
| 2 |
Hb_008948_050 |
0.0707533416 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |
| 3 |
Hb_000093_110 |
0.0714245941 |
- |
- |
PREDICTED: cytochrome b5 isoform A [Jatropha curcas] |
| 4 |
Hb_002588_010 |
0.0785554979 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 5 |
Hb_000062_390 |
0.0813147344 |
- |
- |
PREDICTED: protein Iojap-related, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000567_410 |
0.0888777149 |
- |
- |
hypothetical protein JCGZ_12784 [Jatropha curcas] |
| 7 |
Hb_019516_100 |
0.0921670651 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 8 |
Hb_002909_130 |
0.0930786851 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_116349_110 |
0.0950923376 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 10 |
Hb_002811_080 |
0.0957566521 |
- |
- |
hypothetical protein POPTR_0001s08090g [Populus trichocarpa] |
| 11 |
Hb_000042_260 |
0.0963998346 |
- |
- |
PREDICTED: uncharacterized protein LOC105632814 [Jatropha curcas] |
| 12 |
Hb_001634_040 |
0.0971313888 |
transcription factor |
TF Family: NF-YC |
DNA polymerase epsilon subunit, putative [Ricinus communis] |
| 13 |
Hb_000330_070 |
0.0973685407 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 14 |
Hb_000111_100 |
0.0985432894 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 15 |
Hb_027472_160 |
0.0999884714 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 16 |
Hb_000735_040 |
0.1019383784 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 17 |
Hb_001085_270 |
0.1030597873 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 18 |
Hb_006478_160 |
0.1049771515 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 19 |
Hb_009880_010 |
0.1058156096 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 20 |
Hb_000193_120 |
0.1065006333 |
- |
- |
PREDICTED: protein preY, mitochondrial [Jatropha curcas] |