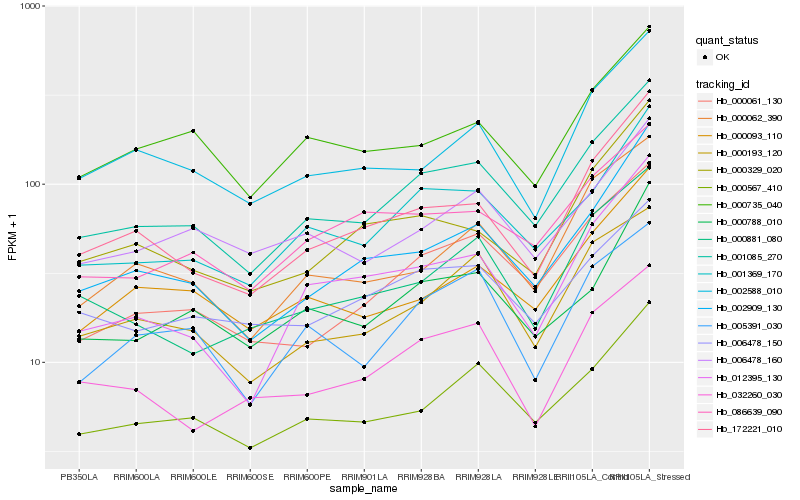
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_270 |
0.0 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 2 |
Hb_001369_170 |
0.059914411 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1-like [Gossypium raimondii] |
| 3 |
Hb_000567_410 |
0.0692152797 |
- |
- |
hypothetical protein JCGZ_12784 [Jatropha curcas] |
| 4 |
Hb_000061_130 |
0.0719506641 |
- |
- |
rotamase, putative [Ricinus communis] |
| 5 |
Hb_002909_130 |
0.0774413309 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_172221_010 |
0.0783484186 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 7 |
Hb_000193_120 |
0.0807370231 |
- |
- |
PREDICTED: protein preY, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000062_390 |
0.0851995483 |
- |
- |
PREDICTED: protein Iojap-related, mitochondrial [Jatropha curcas] |
| 9 |
Hb_086639_090 |
0.0856686371 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 10 |
Hb_000093_110 |
0.0879044473 |
- |
- |
PREDICTED: cytochrome b5 isoform A [Jatropha curcas] |
| 11 |
Hb_012395_130 |
0.0898255037 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 12 |
Hb_000881_080 |
0.0902423676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005391_030 |
0.0911498619 |
- |
- |
putative cytidine/deoxycytidylate deaminase family protein [Zea mays] |
| 14 |
Hb_006478_160 |
0.0918327689 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 15 |
Hb_006478_150 |
0.0920665896 |
- |
- |
mitochondrial ribosomal protein L24, putative [Ricinus communis] |
| 16 |
Hb_000788_010 |
0.0925646753 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 17 |
Hb_000735_040 |
0.0949701507 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 18 |
Hb_002588_010 |
0.0951342908 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 19 |
Hb_032260_030 |
0.0954838664 |
- |
- |
hypothetical protein POPTR_0001s10500g [Populus trichocarpa] |
| 20 |
Hb_000329_020 |
0.0968472572 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |