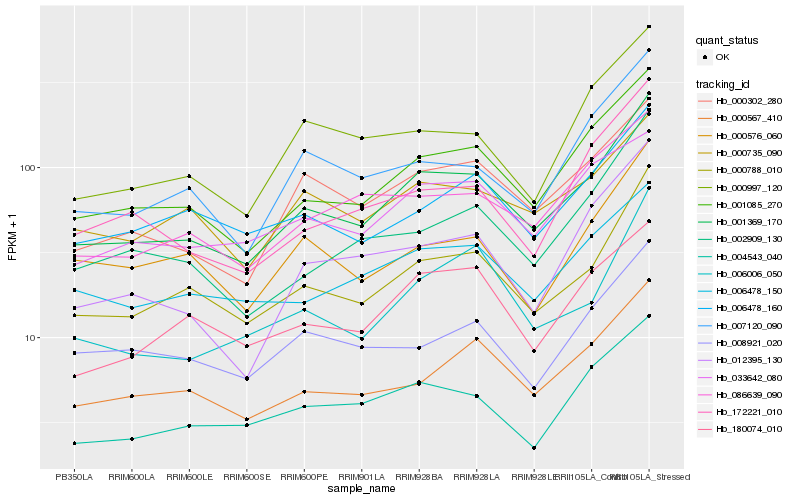
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_170 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1-like [Gossypium raimondii] |
| 2 |
Hb_001085_270 |
0.059914411 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 3 |
Hb_000788_010 |
0.0606855078 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 4 |
Hb_000302_280 |
0.0799284354 |
- |
- |
hypothetical protein ZEAMMB73_772347 [Zea mays] |
| 5 |
Hb_000567_410 |
0.0879675579 |
- |
- |
hypothetical protein JCGZ_12784 [Jatropha curcas] |
| 6 |
Hb_004543_040 |
0.0903273768 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_006478_150 |
0.0932112007 |
- |
- |
mitochondrial ribosomal protein L24, putative [Ricinus communis] |
| 8 |
Hb_000735_090 |
0.0943991134 |
- |
- |
PREDICTED: 40S ribosomal protein S8-like [Gossypium raimondii] |
| 9 |
Hb_086639_090 |
0.0952000466 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 10 |
Hb_006478_160 |
0.096139503 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 11 |
Hb_180074_010 |
0.0977144906 |
- |
- |
Single-stranded DNA-binding protein, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_006006_050 |
0.098966674 |
- |
- |
PREDICTED: methylthioribose kinase-like [Gossypium raimondii] |
| 13 |
Hb_002909_130 |
0.1011922925 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_033642_080 |
0.1025177501 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 15 |
Hb_007120_090 |
0.1026770266 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 16 |
Hb_000576_060 |
0.1048230596 |
- |
- |
truncated hemoglobin [Populus tremula x Populus tremuloides] |
| 17 |
Hb_008921_020 |
0.1059564415 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 1 homolog [Jatropha curcas] |
| 18 |
Hb_000997_120 |
0.1061734744 |
- |
- |
40S ribosomal protein S15E [Hevea brasiliensis] |
| 19 |
Hb_012395_130 |
0.1061746538 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 20 |
Hb_172221_010 |
0.1064093685 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |