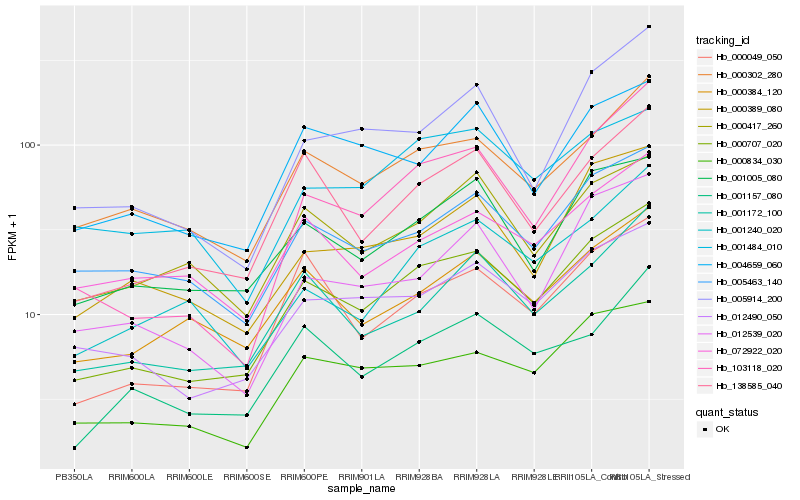
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000707_020 |
0.0 |
- |
- |
hypothetical protein B456_002G255500 [Gossypium raimondii] |
| 2 |
Hb_012490_050 |
0.0863018849 |
- |
- |
PREDICTED: uncharacterized protein LOC105637921 [Jatropha curcas] |
| 3 |
Hb_000834_030 |
0.0917962664 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000302_280 |
0.0954597211 |
- |
- |
hypothetical protein ZEAMMB73_772347 [Zea mays] |
| 5 |
Hb_138585_040 |
0.0962855295 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 6 |
Hb_001005_080 |
0.0979243053 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_000049_050 |
0.0985002436 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 8 |
Hb_103118_020 |
0.1000963687 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 9 |
Hb_001172_100 |
0.1014269055 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001157_080 |
0.1053954445 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000389_080 |
0.1065253424 |
- |
- |
PREDICTED: uncharacterized protein At4g26485-like [Jatropha curcas] |
| 12 |
Hb_005463_140 |
0.1074077109 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18 [Jatropha curcas] |
| 13 |
Hb_005914_200 |
0.1093029147 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 14 |
Hb_000384_120 |
0.1100295734 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000417_260 |
0.1122416332 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_001240_020 |
0.1135983339 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 17 |
Hb_012539_020 |
0.1148101159 |
- |
- |
protein binding/ubiquitin-protein ligase 4 [Hevea brasiliensis] |
| 18 |
Hb_004659_060 |
0.116330749 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 19 |
Hb_001484_010 |
0.1181305057 |
- |
- |
hypothetical protein POPTR_0005s07440g [Populus trichocarpa] |
| 20 |
Hb_072922_020 |
0.1207849431 |
- |
- |
SNARE-like superfamily protein [Theobroma cacao] |