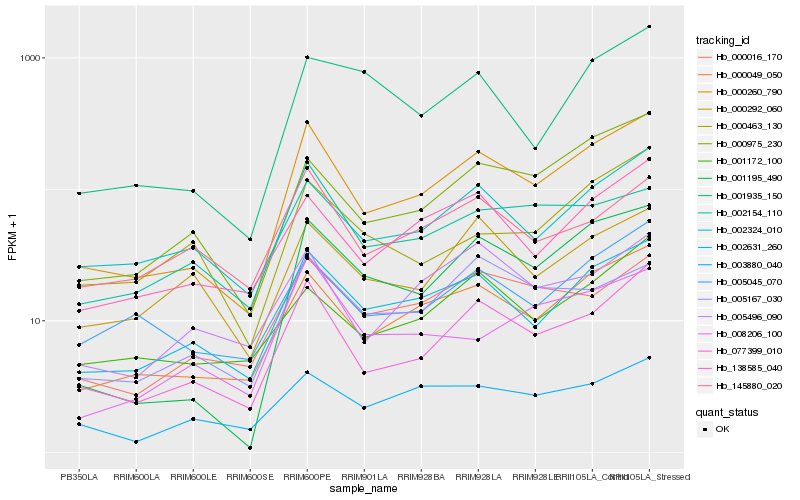
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_790 |
0.0 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 12 isoform 1 [Theobroma cacao] |
| 2 |
Hb_077399_010 |
0.0767771073 |
- |
- |
hypothetical protein POPTR_0003s09910g [Populus trichocarpa] |
| 3 |
Hb_000049_050 |
0.0894360011 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 4 |
Hb_002631_260 |
0.0970176506 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 5 |
Hb_001195_490 |
0.1153037256 |
- |
- |
hypothetical protein glysoja_018593 [Glycine soja] |
| 6 |
Hb_000975_230 |
0.1223214536 |
- |
- |
hypothetical protein ZEAMMB73_864543 [Zea mays] |
| 7 |
Hb_138585_040 |
0.1308865679 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 8 |
Hb_005045_070 |
0.1364069941 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 9 |
Hb_002324_010 |
0.1368354036 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 10 |
Hb_000016_170 |
0.1387911106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005496_090 |
0.1469446601 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 12 |
Hb_005167_030 |
0.1482854296 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 13 |
Hb_001172_100 |
0.1498723684 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_008206_100 |
0.1540450992 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 15 |
Hb_003880_040 |
0.1548352163 |
- |
- |
hypothetical protein POPTR_0001s30770g [Populus trichocarpa] |
| 16 |
Hb_002154_110 |
0.1549660485 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 17 |
Hb_001935_150 |
0.1557333124 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 18 |
Hb_000292_060 |
0.1558904588 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_000463_130 |
0.1624064172 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_145880_020 |
0.1630571709 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |