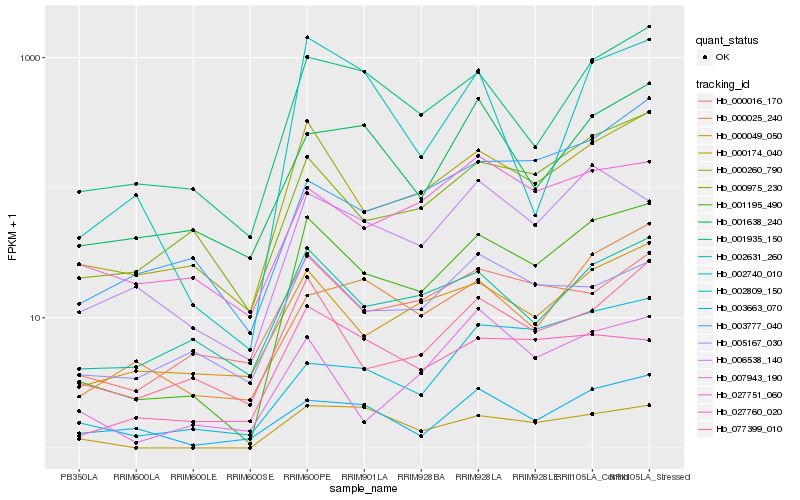
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_490 |
0.0 |
- |
- |
hypothetical protein glysoja_018593 [Glycine soja] |
| 2 |
Hb_000260_790 |
0.1153037256 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 12 isoform 1 [Theobroma cacao] |
| 3 |
Hb_001935_150 |
0.1587358538 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 4 |
Hb_000049_050 |
0.1589509859 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 5 |
Hb_000975_230 |
0.1593794641 |
- |
- |
hypothetical protein ZEAMMB73_864543 [Zea mays] |
| 6 |
Hb_002631_260 |
0.1710625454 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 7 |
Hb_077399_010 |
0.1713087804 |
- |
- |
hypothetical protein POPTR_0003s09910g [Populus trichocarpa] |
| 8 |
Hb_000174_040 |
0.1832636458 |
- |
- |
- |
| 9 |
Hb_002809_150 |
0.1847951494 |
transcription factor |
TF Family: M-type |
hypothetical protein CISIN_1g0283632mg, partial [Citrus sinensis] |
| 10 |
Hb_006538_140 |
0.185111907 |
- |
- |
PREDICTED: uncharacterized protein LOC103341483 [Prunus mume] |
| 11 |
Hb_005167_030 |
0.1906175786 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like, partial [Camelina sativa] |
| 12 |
Hb_003777_040 |
0.1933687207 |
- |
- |
PREDICTED: sm-like protein LSM3A [Jatropha curcas] |
| 13 |
Hb_000025_240 |
0.1938892966 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 14 |
Hb_000016_170 |
0.1941336714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002740_010 |
0.1947042384 |
- |
- |
stress-induced hydrophobic peptide [Cleistogenes songorica] |
| 16 |
Hb_001638_240 |
0.1958101312 |
- |
- |
PREDICTED: uncharacterized protein LOC100262493 [Vitis vinifera] |
| 17 |
Hb_003663_070 |
0.1966574303 |
- |
- |
- |
| 18 |
Hb_007943_190 |
0.1976643545 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 19 |
Hb_027751_060 |
0.1999608319 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 20 |
Hb_027760_020 |
0.2001890268 |
- |
- |
hypothetical protein M569_04462, partial [Genlisea aurea] |